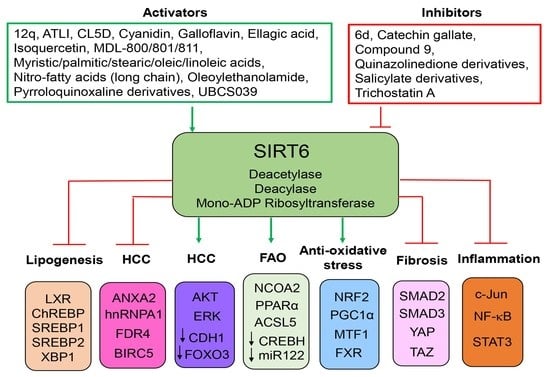
Sirtuin 6—A Key Regulator of Hepatic Lipid Metabolism and Liver Health
Abstract
:1. Introduction
2. SIRT6 in Hepatic Lipogenesis
3. SIRT6 in Hepatic Fatty Acid Oxidation (FAO)
4. SIRT6 in Acute Liver Injury
5. SIRT6 in Hepatic Inflammation
6. SIRT6 in Hepatic Fibrosis
7. SIRT6 in Liver Cancer
8. SIRT6 Chemical Modulators
9. Concluding Remarks
Funding
Data Availability Statement
Conflicts of Interest
References
- Marmorstein, R. Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases. Biochem. Soc. Trans. 2004, 32, 904–909. [Google Scholar] [CrossRef] [Green Version]
- Mahlknecht, U.; Ho, A.D.; Voelter-Mahlknecht, S. Chromosomal organization and fluorescence in situ hybridization of the human Sirtuin 6 gene. Int. J. Oncol. 2006, 28, 447–456. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Xiao, C.; Wang, R.H.; Lahusen, T.; Xu, X.; Vassilopoulos, A.; Vazquez-Ortiz, G.; Jeong, W.I.; Park, O.; Ki, S.H.; et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010, 12, 224–236. [Google Scholar] [CrossRef] [Green Version]
- Tennen, R.I.; Berber, E.; Chua, K.F. Functional dissection of SIRT6: Identification of domains that regulate histone deacetylase activity and chromatin localization. Mech. Ageing Dev. 2010, 131, 185–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liszt, G.; Ford, E.; Kurtev, M.; Guarente, L. Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J. Biol. Chem. 2005, 280, 21313–21320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michishita, E.; McCord, R.A.; Berber, E.; Kioi, M.; Padilla-Nash, H.; Damian, M.; Cheung, P.; Kusumoto, R.; Kawahara, T.L.; Barrett, J.C.; et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452, 492–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michishita, E.; McCord, R.A.; Boxer, L.D.; Barber, M.F.; Hong, T.; Gozani, O.; Chua, K.F. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009, 8, 2664–2666. [Google Scholar] [CrossRef] [Green Version]
- Yang, B.; Zwaans, B.M.; Eckersdorff, M.; Lombard, D.B. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 2009, 8, 2662–2663. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Khan, S.; Wang, Y.; Charron, G.; He, B.; Sebastian, C.; Du, J.; Kim, R.; Ge, E.; Mostoslavsky, R.; et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Spiegelman, N.A.; Nelson, O.D.; Jing, H.; Lin, H. SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. Elife 2017, 6, e25158. [Google Scholar] [CrossRef]
- Zhong, X.; Huang, M.; Kim, H.G.; Zhang, Y.; Chowdhury, K.; Cai, W.; Saxena, R.; Schwabe, R.F.; Liangpunsakul, S.; Dong, X.C. SIRT6 Protects Against Liver Fibrosis by Deacetylation and Suppression of SMAD3 in Hepatic Stellate Cells. Cell Mol. Gastroenterol. Hepatol. 2020, 10, 341–364. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, K.; Huang, M.; Kim, H.G.; Dong, X.C. Sirtuin 6 protects against hepatic fibrogenesis by suppressing the YAP and TAZ function. FASEB J. 2022, 36, e22529. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Liu, Q.; Huang, Y.; Li, R.; Wu, T.; Zhang, Z.; Zhou, J.; Huang, H.; Tang, Q.; et al. Sirt6 Alleviated Liver Fibrosis by Deacetylating Conserved Lysine 54 on Smad2 in Hepatic Stellate Cells. Hepatology 2021, 73, 1140–1157. [Google Scholar] [CrossRef]
- Dominy, J.E., Jr.; Lee, Y.; Jedrychowski, M.P.; Chim, H.; Jurczak, M.J.; Camporez, J.P.; Ruan, H.B.; Feldman, J.; Pierce, K.; Mostoslavsky, R.; et al. The deacetylase Sirt6 activates the acetyltransferase GCN5 and suppresses hepatic gluconeogenesis. Mol. Cell 2012, 48, 900–913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bang, I.H.; Kwon, O.K.; Hao, L.; Park, D.; Chung, M.J.; Oh, B.C.; Lee, S.; Bae, E.J.; Park, B.H. Deacetylation of XBP1s by sirtuin 6 confers resistance to ER stress-induced hepatic steatosis. Exp. Mol. Med. 2019, 51, 107. [Google Scholar] [CrossRef]
- Hou, T.; Tian, Y.; Cao, Z.; Zhang, J.; Feng, T.; Tao, W.; Sun, H.; Wen, H.; Lu, X.; Zhu, Q.; et al. Cytoplasmic SIRT6-mediated ACSL5 deacetylation impedes nonalcoholic fatty liver disease by facilitating hepatic fatty acid oxidation. Mol. Cell 2022, 82, 4099–4115.e4099. [Google Scholar] [CrossRef]
- Han, L.L.; Jia, L.; Wu, F.; Huang, C. Sirtuin6 (SIRT6) Promotes the EMT of Hepatocellular Carcinoma by Stimulating Autophagic Degradation of E-Cadherin. Mol. Cancer Res. 2019, 17, 2267–2280. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Jia, X.; Yang, X.; Bai, X.; Lu, Y.; Zhu, L.; Cheng, W.; Shu, M.; Zhu, Y.; Du, X.; et al. Deacetylation of Caveolin-1 by Sirt6 induces autophagy and retards high glucose-stimulated LDL transcytosis and atherosclerosis formation. Metabolism 2022, 131, 155162. [Google Scholar] [CrossRef]
- Wang, H.; Feng, K.; Wang, Q.; Deng, H. Reciprocal interaction between SIRT6 and APC/C regulates genomic stability. Sci. Rep. 2021, 11, 14253. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Huang, M.; Kim, H.G.; Chowdhury, K.; Gao, J.; Liu, S.; Wan, J.; Wei, L.; Dong, X.C. SIRT6 controls hepatic lipogenesis by suppressing LXR, ChREBP, and SREBP1. Biochim. Biophys Acta Mol. Basis. Dis. 2021, 1867, 166249. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Li, W.; Wang, T.; Park, B.H.; Park, S.K.; Kang, K.P. Loss of Proximal Tubular Sirtuin 6 Aggravates Unilateral Ureteral Obstruction-Induced Tubulointerstitial Inflammation and Fibrosis by Regulation of beta-Catenin Acetylation. Cells 2022, 11, 1477. [Google Scholar] [CrossRef] [PubMed]
- Geng, A.; Tang, H.; Huang, J.; Qian, Z.; Qin, N.; Yao, Y.; Xu, Z.; Chen, H.; Lan, L.; Xie, H.; et al. The deacetylase SIRT6 promotes the repair of UV-induced DNA damage by targeting DDB2. Nucleic Acids Res. 2020, 48, 9181–9194. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Chen, J.; Wang, Q.; Sun, X.; Han, J.; Guastaldi, F.; Xiang, S.; Ye, Q.; He, Y. SIRT6 Promotes Osteogenic Differentiation of Adipose-Derived Mesenchymal Stem Cells through Antagonizing DNMT1. Front. Cell Dev. Biol. 2021, 9, 648627. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, S.; Li, B.; Li, Y.; Aman, S.; Xia, K.; Yang, Y.; Ahmad, B.; Wu, H. Acetylation of ELF5 suppresses breast cancer progression by promoting its degradation and targeting CCND1. NPJ Precis. Oncol. 2021, 5, 20. [Google Scholar] [CrossRef] [PubMed]
- Moon, Y.J.; Zhang, Z.; Bang, I.H.; Kwon, O.K.; Yoon, S.J.; Kim, J.R.; Lee, S.; Bae, E.J.; Park, B.H. Sirtuin 6 in preosteoclasts suppresses age- and estrogen deficiency-related bone loss by stabilizing estrogen receptor alpha. Cell Death Differ. 2019, 26, 2358–2370. [Google Scholar] [CrossRef]
- Hao, L.; Bang, I.H.; Wang, J.; Mao, Y.; Yang, J.D.; Na, S.Y.; Seo, J.K.; Choi, H.S.; Bae, E.J.; Park, B.H. ERRgamma suppression by Sirt6 alleviates cholestatic liver injury and fibrosis. JCI Insight 2020, 5, e137566. [Google Scholar] [CrossRef] [PubMed]
- He, T.; Shang, J.; Gao, C.; Guan, X.; Chen, Y.; Zhu, L.; Zhang, L.; Zhang, C.; Zhang, J.; Pang, T. A novel SIRT6 activator ameliorates neuroinflammation and ischemic brain injury via EZH2/FOXC1 axis. Acta Pharm. Sin. B 2021, 11, 708–726. [Google Scholar] [CrossRef]
- Liu, T.; Li, Z.; Tian, F. Quercetin inhibited the proliferation and invasion of hepatoblastoma cells through facilitating SIRT6-medicated FZD4 silence. Hum. Exp. Toxicol. 2021, 40, S96–S107. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Jiang, W.; Liu, M.; Wang, Y.; Ma, H.; Mu, N.; Wang, H. SIRT6 Protects Against Myocardial Ischemia-Reperfusion Injury by Attenuating Aging-Related CHMP2B Accumulation. J. Cardiovasc Transl. Res. 2022, 15, 740–753. [Google Scholar] [CrossRef]
- Jung, S.M.; Hung, C.M.; Hildebrand, S.R.; Sanchez-Gurmaches, J.; Martinez-Pastor, B.; Gengatharan, J.M.; Wallace, M.; Mukhopadhyay, D.; Martinez Calejman, C.; Luciano, A.K.; et al. Non-canonical mTORC2 Signaling Regulates Brown Adipocyte Lipid Catabolism through SIRT6-FoxO1. Mol. Cell 2019, 75, 807–822.e808. [Google Scholar] [CrossRef]
- Khongkow, M.; Olmos, Y.; Gong, C.; Gomes, A.R.; Monteiro, L.J.; Yague, E.; Cavaco, T.B.; Khongkow, P.; Man, E.P.; Laohasinnarong, S.; et al. SIRT6 modulates paclitaxel and epirubicin resistance and survival in breast cancer. Carcinogenesis 2013, 34, 1476–1486. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Zhang, Y.; Liu, Q.; Shen, J.; Pu, S.; Cheng, S.; Chen, L.; Li, H.; Wu, T.; Li, R.; et al. Fat-Specific Sirt6 Ablation Sensitizes Mice to High-Fat Diet-Induced Obesity and Insulin Resistance by Inhibiting Lipolysis. Diabetes 2017, 66, 1159–1171. [Google Scholar] [CrossRef] [Green Version]
- Shi, M.Y.; Bang, I.H.; Han, C.Y.; Lee, D.H.; Park, B.H.; Bae, E.J. Statin suppresses sirtuin 6 through miR-495, increasing FoxO1-dependent hepatic gluconeogenesis. Theranostics 2020, 10, 11416–11427. [Google Scholar] [CrossRef]
- Song, M.Y.; Wang, J.; Ka, S.O.; Bae, E.J.; Park, B.H. Insulin secretion impairment in Sirt6 knockout pancreatic beta cells is mediated by suppression of the FoxO1-Pdx1-Glut2 pathway. Sci. Rep. 2016, 6, 30321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, S.J.; Noh, H.S.; Lee, N.Y.; Cheon, Y.H.; Yi, S.M.; Jeon, H.M.; Bae, E.J.; Lee, S.I.; Park, B.H. Myeloid sirtuin 6 deficiency accelerates experimental rheumatoid arthritis by enhancing macrophage activation and infiltration into synovium. EBioMedicine 2018, 38, 228–237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Tu, B.; Wang, H.; Cao, Z.; Tang, M.; Zhang, C.; Gu, B.; Li, Z.; Wang, L.; Yang, Y.; et al. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proc. Natl. Acad. Sci. USA 2014, 111, 10684–10689. [Google Scholar] [CrossRef] [Green Version]
- Mahmud, Z.; Gomes, A.R.; Lee, H.J.; Aimjongjun, S.; Jiramongkol, Y.; Yao, S.; Zona, S.; Alasiri, G.; Gong, G.; Yague, E.; et al. EP300 and SIRT1/6 Co-Regulate Lapatinib Sensitivity via Modulating FOXO3-Acetylation and Activity in Breast Cancer. Cancers 2019, 11, 1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Pan, Z.; Wu, Z.; Tang, K.; Zhong, Y.; Chen, Y.; Xiao, X.; Guo, J.; Duan, S.; Cui, T.; et al. Hepatic SIRT6 Modulates Transcriptional Activities of FXR to Alleviate Acetaminophen-induced Hepatotoxicity. Cell Mol. Gastroenterol Hepatol. 2022, 14, 271–293. [Google Scholar] [CrossRef]
- Jang, H.Y.; Gu, S.; Lee, S.M.; Park, B.H. Overexpression of sirtuin 6 suppresses allergic airway inflammation through deacetylation of GATA3. J. Allergy Clin. Immunol. 2016, 138, 1452–1455.e1413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, D.; Li, W.; Xie, C.; Yin, L.; Su, X.; Chen, J.; Huang, H. Capsaicin Attenuates Arterial Calcification through Promoting SIRT6-Mediated Deacetylation and Degradation of Hif1alpha (Hypoxic-Inducible Factor-1 Alpha). Hypertension 2022, 79, 906–917. [Google Scholar] [CrossRef]
- Tasselli, L.; Xi, Y.; Zheng, W.; Tennen, R.I.; Odrowaz, Z.; Simeoni, F.; Li, W.; Chua, K.F. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat. Struct. Mol. Biol. 2016, 23, 434–440. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.; Guo, X.; Gu, L.; Jia, J.; Yang, M.; Yuan, W.; Rong, S. Bone marrow mesenchymal stem cell exosomes suppress phosphate-induced aortic calcification via SIRT6-HMGB1 deacetylation. Stem Cell Res. Ther. 2021, 12, 235. [Google Scholar] [CrossRef]
- Yang, H.; Zhu, R.; Zhao, X.; Liu, L.; Zhou, Z.; Zhao, L.; Liang, B.; Ma, W.; Zhao, J.; Liu, J.; et al. Sirtuin-mediated deacetylation of hnRNP A1 suppresses glycolysis and growth in hepatocellular carcinoma. Oncogene 2019, 38, 4915–4931. [Google Scholar] [CrossRef]
- Tao, N.N.; Ren, J.H.; Tang, H.; Ran, L.K.; Zhou, H.Z.; Liu, B.; Huang, A.L.; Chen, J. Deacetylation of Ku70 by SIRT6 attenuates Bax-mediated apoptosis in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2017, 485, 713–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Bai, L.; Ren, Q.; Sun, G.; Si, Y. Protective effects of SIRT6 against lipopolysaccharide (LPS) are mediated by deacetylation of Ku70. Mol. Immunol. 2018, 101, 312–318. [Google Scholar] [CrossRef]
- Huang, S.; Shao, T.; Liu, H.; Wang, Q.; Li, T.; Zhao, Q. SIRT6 mediates MRTF-A deacetylation in vascular endothelial cells to antagonize oxLDL-induced ICAM-1 transcription. Cell Death Discov. 2022, 8, 96. [Google Scholar] [CrossRef]
- Sociali, G.; Grozio, A.; Caffa, I.; Schuster, S.; Becherini, P.; Damonte, P.; Sturla, L.; Fresia, C.; Passalacqua, M.; Mazzola, F.; et al. SIRT6 deacetylase activity regulates NAMPT activity and NAD(P)(H) pools in cancer cells. FASEB J. 2019, 33, 3704–3717. [Google Scholar] [CrossRef]
- Naiman, S.; Huynh, F.K.; Gil, R.; Glick, Y.; Shahar, Y.; Touitou, N.; Nahum, L.; Avivi, M.Y.; Roichman, A.; Kanfi, Y.; et al. SIRT6 Promotes Hepatic Beta-Oxidation via Activation of PPARalpha. Cell Rep. 2019, 29, 4127–4143.e4128. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhang, X.; Guo, Z.; Zhong, Y.; Wang, P.; Li, J.; Li, Z.; Liu, P. SIRT6 Suppresses NFATc4 Expression and Activation in Cardiomyocyte Hypertrophy. Front. Pharmacol. 2018, 9, 1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Jiang, D.; Huang, W.; Teng, P.; Zhang, H.; Wei, C.; Cai, X.; Liang, Y. Sirtuin 6 attenuates angiotensin II-induced vascular adventitial aging in rat aortae by suppressing the NF-kappaB pathway. Hypertens. Res. 2021, 44, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Wu, Y.; Fu, D.; Liu, Y.; Huang, C. SIRT6 regulates osteogenic differentiation of rat bone marrow mesenchymal stem cells partially via suppressing the nuclear factor-kappaB signaling pathway. Stem Cells 2014, 32, 1943–1955. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Liu, Z.; Lu, J.; Zou, J.; Ye, W.; Li, H.; Gao, S.; Liu, P. SIRT6 regulates endothelium-dependent relaxation by modulating nitric oxide synthase 3 (NOS3). Biochem. Pharmacol. 2023, 209, 115439. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Guo, J.; Tang, K.; Chen, Y.; Gong, X.; Chen, Y.; Zhong, Y.; Xiao, X.; Duan, S.; Cui, T.; et al. Ginsenoside Rc Modulates SIRT6-NRF2 Interaction to Alleviate Alcoholic Liver Disease. J. Agric. Food Chem. 2022, 70, 14220–14234. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Wong, S.K.; Jiang, Z.; Liu, B.; Wang, Y.; Hao, Q.; Gorbunova, V.; Liu, X.; Zhou, Z. Haploinsufficiency of Trp53 dramatically extends the lifespan of Sirt6-deficient mice. Elife 2018, 7, e32127. [Google Scholar] [CrossRef]
- Sun, S.; Liu, Z.; Feng, Y.; Shi, L.; Cao, X.; Cai, Y.; Liu, B. Sirt6 deacetylase activity regulates circadian rhythms via Per2. Biochem. Biophys. Res. Commun. 2019, 511, 234–238. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Das, S. SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc. Natl. Acad. Sci. USA 2016, 113, E538–E547. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sheng, Z.; Dong, Z.; Wu, Q.; Cai, Y. The mechanism of radiotherapy for lung adenocarcinoma in promoting protein SIRT6-mediated deacetylation of RBBP8 to enhance the sensitivity of targeted therapy. Int. J. Immunopathol. Pharmacol. 2022, 36, 3946320221130727. [Google Scholar] [CrossRef]
- Li, W.; Feng, W.; Su, X.; Luo, D.; Li, Z.; Zhou, Y.; Zhu, Y.; Zhang, M.; Chen, J.; Liu, B.; et al. SIRT6 protects vascular smooth muscle cells from osteogenic transdifferentiation via Runx2 in chronic kidney disease. J. Clin. Investig. 2022, 132, e150051. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Liu, Q.; Yang, X.; Wu, T.; Huang, C.; Zhang, J.; Zhang, Z.; Zhang, G.; Zhao, Y.; Zhou, J.; et al. Sirtuin 6 supra-physiological overexpression in hypothalamic pro-opiomelanocortin neurons promotes obesity via the hypothalamus-adipose axis. FASEB J. 2021, 35, e21408. [Google Scholar] [CrossRef]
- Ji, M.L.; Jiang, H.; Li, Z.; Geng, R.; Hu, J.Z.; Lin, Y.C.; Lu, J. Sirt6 attenuates chondrocyte senescence and osteoarthritis progression. Nat. Commun. 2022, 13, 7658. [Google Scholar] [CrossRef] [PubMed]
- Portillo, M.; Eremenko, E.; Kaluski, S.; Garcia-Venzor, A.; Onn, L.; Stein, D.; Slobodnik, Z.; Zaretsky, A.; Ueberham, U.; Einav, M.; et al. SIRT6-CBP-dependent nuclear Tau accumulation and its role in protein synthesis. Cell Rep. 2021, 35, 109035. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, A.; Iachettini, S.; Salvati, E.; Zizza, P.; Maresca, C.; D’Angelo, C.; Benarroch-Popivker, D.; Capolupo, A.; Del Gaudio, F.; Cosconati, S.; et al. SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res. 2017, 45, 1820–1834. [Google Scholar] [CrossRef] [PubMed]
- Rezazadeh, S.; Yang, D.; Tombline, G.; Simon, M.; Regan, S.P.; Seluanov, A.; Gorbunova, V. SIRT6 promotes transcription of a subset of NRF2 targets by mono-ADP-ribosylating BAF170. Nucleic Acids Res. 2019, 47, 7914–7928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Meter, M.; Kashyap, M.; Rezazadeh, S.; Geneva, A.J.; Morello, T.D.; Seluanov, A.; Gorbunova, V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014, 5, 5011. [Google Scholar] [CrossRef] [Green Version]
- Rezazadeh, S.; Yang, D.; Biashad, S.A.; Firsanov, D.; Takasugi, M.; Gilbert, M.; Tombline, G.; Bhanu, N.V.; Garcia, B.A.; Seluanov, A.; et al. SIRT6 mono-ADP ribosylates KDM2A to locally increase H3K36me2 at DNA damage sites to inhibit transcription and promote repair. Aging 2020, 12, 11165–11184. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Yang, J.; Gigas, J.; Earley, E.J.; Hillpot, E.; Zhang, L.; Zagorulya, M.; Tombline, G.; Gilbert, M.; Yuen, S.L.; et al. A rare human centenarian variant of SIRT6 enhances genome stability and interaction with Lamin A. EMBO J. 2022, 41, e110393. [Google Scholar] [CrossRef]
- Mao, Z.; Hine, C.; Tian, X.; Van Meter, M.; Au, M.; Vaidya, A.; Seluanov, A.; Gorbunova, V. SIRT6 promotes DNA repair under stress by activating PARP1. Science 2011, 332, 1443–1446. [Google Scholar] [CrossRef] [Green Version]
- Mostoslavsky, R.; Chua, K.F.; Lombard, D.B.; Pang, W.W.; Fischer, M.R.; Gellon, L.; Liu, P.; Mostoslavsky, G.; Franco, S.; Murphy, M.M.; et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 2006, 124, 315–329. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Wan, H.; Feng, G.; Qu, J.; Wang, J.; Jing, Y.; Ren, R.; Liu, Z.; Zhang, L.; Chen, Z.; et al. SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 2018, 560, 661–665. [Google Scholar] [CrossRef]
- Ferrer, C.M.; Alders, M.; Postma, A.V.; Park, S.; Klein, M.A.; Cetinbas, M.; Pajkrt, E.; Glas, A.; van Koningsbruggen, S.; Christoffels, V.M.; et al. An inactivating mutation in the histone deacetylase SIRT6 causes human perinatal lethality. Genes Dev. 2018, 32, 373–388. [Google Scholar] [CrossRef] [Green Version]
- Kugel, S.; Feldman, J.L.; Klein, M.A.; Silberman, D.M.; Sebastian, C.; Mermel, C.; Dobersch, S.; Clark, A.R.; Getz, G.; Denu, J.M.; et al. Identification of and Molecular Basis for SIRT6 Loss-of-Function Point Mutations in Cancer. Cell Rep. 2015, 13, 479–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masri, S.; Rigor, P.; Cervantes, M.; Ceglia, N.; Sebastian, C.; Xiao, C.; Roqueta-Rivera, M.; Deng, C.; Osborne, T.F.; Mostoslavsky, R.; et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell 2014, 158, 659–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ka, S.O.; Bang, I.H.; Bae, E.J.; Park, B.H. Hepatocyte-specific sirtuin 6 deletion predisposes to nonalcoholic steatohepatitis by up-regulation of Bach1, an Nrf2 repressor. FASEB J. 2017, 31, 3999–4010. [Google Scholar] [CrossRef]
- Luo, P.; Qin, C.; Zhu, L.; Fang, C.; Zhang, Y.; Zhang, H.; Pei, F.; Tian, S.; Zhu, X.Y.; Gong, J.; et al. Ubiquitin-Specific Peptidase 10 (USP10) Inhibits Hepatic Steatosis, Insulin Resistance, and Inflammation through Sirt6. Hepatology 2018, 68, 1786–1803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.X.; Dong, X.C. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J. Lipid Res. 2013, 54, 2745–2753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elhanati, S.; Kanfi, Y.; Varvak, A.; Roichman, A.; Carmel-Gross, I.; Barth, S.; Gibor, G.; Cohen, H.Y. Multiple regulatory layers of SREBP1/2 by SIRT6. Cell Rep. 2013, 4, 905–912. [Google Scholar] [CrossRef] [Green Version]
- Tao, R.; Xiong, X.; DePinho, R.A.; Deng, C.X.; Dong, X.C. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J. Biol. Chem. 2013, 288, 29252–29259. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Liu, Q.; Tang, Q.; Kuang, J.; Li, H.; Pu, S.; Wu, T.; Yang, X.; Li, R.; Zhang, J.; et al. Hepatocyte-specific Sirt6 deficiency impairs ketogenesis. J. Biol. Chem. 2019, 294, 1579–1589. [Google Scholar] [CrossRef] [Green Version]
- Elhanati, S.; Ben-Hamo, R.; Kanfi, Y.; Varvak, A.; Glazz, R.; Lerrer, B.; Efroni, S.; Cohen, H.Y. Reciprocal Regulation between SIRT6 and miR-122 Controls Liver Metabolism and Predicts Hepatocarcinoma Prognosis. Cell Rep. 2016, 14, 234–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.G.; Huang, M.; Xin, Y.; Zhang, Y.; Zhang, X.; Wang, G.; Liu, S.; Wan, J.; Ahmadi, A.R.; Sun, Z.; et al. The epigenetic regulator SIRT6 protects the liver from alcohol-induced tissue injury by reducing oxidative stress in mice. J. Hepatol. 2019, 71, 960–969. [Google Scholar] [CrossRef]
- Xin, Y.; Xu, L.; Zhang, X.; Yang, C.; Wang, Q.; Xiong, X. Sirtuin 6 ameliorates alcohol-induced liver injury by reducing endoplasmic reticulum stress in mice. Biochem. Biophys. Res. Commun. 2021, 544, 44–51. [Google Scholar] [CrossRef]
- Gao, S.; Yang, Q.; Liu, Z.; Kong, W.; Chen, J.; Li, X.; Peng, Y.; Bao, M.; Bian, X.; Zhang, Y.; et al. Metformin alleviates HFD-induced oxidative stress in hepatocyte via activating SIRT6/PGC-1alpha/ENDOG signaling. Clin. Sci. 2022, 136, 1711–1730. [Google Scholar] [CrossRef]
- Zhou, Y.; Fan, X.; Jiao, T.; Li, W.; Chen, P.; Jiang, Y.; Sun, J.; Chen, Y.; Chen, P.; Guan, L.; et al. SIRT6 as a key event linking P53 and NRF2 counteracts APAP-induced hepatotoxicity through inhibiting oxidative stress and promoting hepatocyte proliferation. Acta Pharm. Sin. B 2021, 11, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Jiang, S.; Wang, H.; Di, W.; Deng, C.; Jin, Z.; Yi, W.; Xiao, X.; Nie, Y.; Yang, Y. SIRT6 protects against hepatic ischemia/reperfusion injury by inhibiting apoptosis and autophagy related cell death. Free Radic. Biol. Med. 2018, 115, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wang, R.H.; Lahusen, T.J.; Park, O.; Bertola, A.; Maruyama, T.; Reynolds, D.; Chen, Q.; Xu, X.; Young, H.A.; et al. Progression of Chronic Liver Inflammation and Fibrosis Driven by Activation of c-JUN Signaling in Sirt6 Mutant Mice. J. Biol. Chem. 2012, 287, 41903–41913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Ka, S.O.; Cha, H.N.; Chae, Y.N.; Kim, M.K.; Park, S.Y.; Bae, E.J.; Park, B.H. Myeloid Sirtuin 6 Deficiency Causes Insulin Resistance in High-Fat Diet-Fed Mice by Eliciting Macrophage Polarization toward an M1 Phenotype. Diabetes 2017, 66, 2659–2668. [Google Scholar] [CrossRef] [Green Version]
- Maity, S.; Muhamed, J.; Sarikhani, M.; Kumar, S.; Ahamed, F.; Spurthi, K.M.; Ravi, V.; Jain, A.; Khan, D.; Arathi, B.P.; et al. Sirtuin 6 deficiency transcriptionally up-regulates TGF-beta signaling and induces fibrosis in mice. J. Biol. Chem. 2020, 295, 415–434. [Google Scholar] [CrossRef]
- Xiang, X.; Ohshiro, K.; Zaidi, S.; Yang, X.; Bhowmick, K.; Vegesna, A.K.; Bernstein, D.; Crawford, J.M.; Mishra, B.; Latham, P.S.; et al. Impaired reciprocal regulation between SIRT6 and TGF-beta signaling in fatty liver. FASEB J. 2022, 36, e22335. [Google Scholar] [CrossRef]
- Mitten, E.K.; Baffy, G. Mechanotransduction in the pathogenesis of non-alcoholic fatty liver disease. J. Hepatol. 2022, 77, 1642–1656. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, J.; Raffaele, M.; Skalova, H.; Leire, E.; Pata, I.; Pata, P.; Gorbunova, V.; Vinciguerra, M. Human centenarian-associated SIRT6 mutants modulate hepatocyte metabolism and collagen deposition in multilineage hepatic 3D spheroids. Geroscience 2022, 45, 1177–1196. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lan, L.; Yang, F.; Jiang, S.; Xu, H.; Zhang, C.; Zhou, G.; Xia, H.; Xia, J. Hepatic SIRT6 deficit promotes liver tumorigenesis in the mice models. Genes Dis. 2022, 9, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, Y.; Kang, M.; Feng, M.; Ren, Y.; Dai, H.; Wang, Y.; Wang, Y.; Tang, B. USP48 is upregulated by Mettl14 to attenuate hepatocellular carcinoma via regulating SIRT6 stabilization. Cancer Res. 2021, 81, 3822–3834. [Google Scholar] [CrossRef] [PubMed]
- Kohli, S.; Bhardwaj, A.; Kumari, R.; Das, S. SIRT6 Is a Target of Regulation by UBE3A That Contributes to Liver Tumorigenesis in an ANXA2-Dependent Manner. Cancer Res. 2018, 78, 645–658. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.G.; Qin, C.Y. Sirt6 suppresses hepatocellular carcinoma cell growth via inhibiting the extracellular signalregulated kinase signaling pathway. Mol. Med. Rep. 2014, 9, 882–888. [Google Scholar] [CrossRef] [Green Version]
- Marquardt, J.U.; Fischer, K.; Baus, K.; Kashyap, A.; Ma, S.; Krupp, M.; Linke, M.; Teufel, A.; Zechner, U.; Strand, D.; et al. Sirtuin-6-dependent genetic and epigenetic alterations are associated with poor clinical outcome in hepatocellular carcinoma patients. Hepatology 2013, 58, 1054–1064. [Google Scholar] [CrossRef] [Green Version]
- Min, L.; Ji, Y.; Bakiri, L.; Qiu, Z.; Cen, J.; Chen, X.; Chen, L.; Scheuch, H.; Zheng, H.; Qin, L.; et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat. Cell. Biol. 2012, 14, 1203–1211. [Google Scholar] [CrossRef]
- Han, L.; Jia, L.; Zan, Y. Long intergenic noncoding RNA smad7 (Linc-smad7) promotes the epithelial-mesenchymal transition of HCC by targeting the miR-125b/SIRT6 axis. Cancer Med. 2020, 9, 9123–9137. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.Z.; Zeng, H.Q.; Yuan, D.; Ren, J.H.; Cheng, S.T.; Yu, H.B.; Ren, F.; Wang, Q.; Qin, Y.P.; Huang, A.L.; et al. NQO1 potentiates apoptosis evasion and upregulates XIAP via inhibiting proteasome-mediated degradation SIRT6 in hepatocellular carcinoma. Cell Commun. Signal. 2019, 17, 168. [Google Scholar] [CrossRef] [Green Version]
- Ran, L.K.; Chen, Y.; Zhang, Z.Z.; Tao, N.N.; Ren, J.H.; Zhou, L.; Tang, H.; Chen, X.; Chen, K.; Li, W.Y.; et al. SIRT6 Overexpression Potentiates Apoptosis Evasion in Hepatocellular Carcinoma via BCL2-Associated X Protein-Dependent Apoptotic Pathway. Clin. Cancer Res. 2016, 22, 3372–3382. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Yu, Y.; Huang, Q.; Tang, K. SIRT6 regulates the proliferation and apoptosis of hepatocellular carcinoma via the ERK1/2 signaling pathway. Mol. Med. Rep. 2019, 20, 1575–1582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.Q.; Deng, F.; Hu, X.P.; Zhang, W.; Zeng, X.C.; Tian, X.F. Histone deacetylase SIRT6 regulates chemosensitivity in liver cancer cells via modulation of FOXO3 activity. Oncol. Rep. 2018, 40, 3635–3644. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.X.; Luo, J.; Liu, M.; Yan, W.; Zhou, Z.Z.; Xia, Y.J.; Tu, W.; Li, P.Y.; Feng, Z.H.; Tian, D.A. Sirtuin 6 promotes transforming growth factor-beta1/H2O2/HOCl-mediated enhancement of hepatocellular carcinoma cell tumorigenicity by suppressing cellular senescence. Cancer Sci. 2015, 106, 559–566. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Ryu, H.G.; Kwon, J.H.; Kim, D.K.; Kim, S.R.; Wang, H.J.; Kim, K.T.; Choi, K.Y. SIRT6 Depletion Suppresses Tumor Growth by Promoting Cellular Senescence Induced by DNA Damage in HCC. PLoS ONE 2016, 11, e0165835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Shi, S.; Liu, G.; Xie, X.; Li, J.; Bolinger, A.A.; Chen, H.; Zhang, W.; Shi, P.Y.; Liu, H.; et al. Design, synthesis, and pharmacological evaluations of pyrrolo[1,2-a]quinoxaline-based derivatives as potent and selective sirt6 activators. Eur. J. Med. Chem. 2023, 246, 114998. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Mai, Z.; Chen, Y.; Luo, L.; Liu, H.; Zhao, L.; Huang, R.; Wang, S.; Chen, R.; Zhou, H.; et al. ATL I, Acts as a SIRT6 Activator to Alleviate Hepatic Steatosis in Mice via Suppression of NLRP3 Inflammasome Formation. Pharmaceuticals 2022, 15, 1526. [Google Scholar] [CrossRef] [PubMed]
- Carreno, M.; Bresque, M.; Machado, M.R.; Santos, L.; Duran, R.; Vitturi, D.A.; Escande, C.; Denicola, A. Nitro-fatty acids as activators of hSIRT6 deacetylase activity. J. Biol. Chem. 2020, 295, 18355–18366. [Google Scholar] [CrossRef]
- Rahnasto-Rilla, M.; Tyni, J.; Huovinen, M.; Jarho, E.; Kulikowicz, T.; Ravichandran, S.; Vilhelm, A.B.; Ferrucci, L.; Lahtela-Kakkonen, M.; Moaddel, R. Natural polyphenols as sirtuin 6 modulators. Sci. Rep. 2018, 8, 4163. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Sun, W.; Huang, S.; Zhang, H.; Lin, G.; Li, H.; Qiao, J.; Li, L.; Yang, S. Discovery of Potent Small-Molecule SIRT6 Activators: Structure-Activity Relationship and Anti-Pancreatic Ductal Adenocarcinoma Activity. J. Med. Chem. 2020, 63, 10474–10495. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Zhao, J.; Deng, W.; Chen, Y.; Shang, J.; Song, K.; Zhang, L.; Wang, C.; Lu, S.; Yang, X.; et al. Identification of a cellularly active SIRT6 allosteric activator. Nat. Chem. Biol. 2018, 14, 1118–1126. [Google Scholar] [CrossRef]
- Shang, J.; Zhu, Z.; Chen, Y.; Song, J.; Huang, Y.; Song, K.; Zhong, J.; Xu, X.; Wei, J.; Wang, C.; et al. Small-molecule activating SIRT6 elicits therapeutic effects and synergistically promotes anti-tumor activity of vitamin D(3) in colorectal cancer. Theranostics 2020, 10, 5845–5864. [Google Scholar] [CrossRef]
- Feldman, J.L.; Baeza, J.; Denu, J.M. Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J. Biol. Chem. 2013, 288, 31350–31356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahnasto-Rilla, M.; Kokkola, T.; Jarho, E.; Lahtela-Kakkonen, M.; Moaddel, R. N-Acylethanolamines Bind to SIRT6. Chembiochem 2016, 17, 77–81. [Google Scholar] [CrossRef] [Green Version]
- Klein, M.A.; Denu, J.M. Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators. J. Biol. Chem. 2020, 295, 11021–11041. [Google Scholar] [CrossRef] [PubMed]
- You, W.; Zheng, W.; Weiss, S.; Chua, K.F.; Steegborn, C. Structural basis for the activation and inhibition of Sirtuin 6 by quercetin and its derivatives. Sci. Rep. 2019, 9, 19176. [Google Scholar] [CrossRef] [Green Version]
- You, W.; Rotili, D.; Li, T.M.; Kambach, C.; Meleshin, M.; Schutkowski, M.; Chua, K.F.; Mai, A.; Steegborn, C. Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules. Angew. Chem. Int. Ed. Engl. 2017, 56, 1007–1011. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Chen, X.; Huang, S.; Li, W.; Tian, C.; Yang, S.; Li, L. Discovery of 5-(4-methylpiperazin-1-yl)-2-nitroaniline derivatives as a new class of SIRT6 inhibitors. Bioorg. Med. Chem. Lett. 2020, 30, 127215. [Google Scholar] [CrossRef] [PubMed]
- Wood, M.; Rymarchyk, S.; Zheng, S.; Cen, Y. Trichostatin A inhibits deacetylation of histone H3 and p53 by SIRT6. Arch Biochem. Biophys. 2018, 638, 8–17. [Google Scholar] [CrossRef] [PubMed]
- Damonte, P.; Sociali, G.; Parenti, M.D.; Soncini, D.; Bauer, I.; Boero, S.; Grozio, A.; Holtey, M.V.; Piacente, F.; Becherini, P.; et al. SIRT6 inhibitors with salicylate-like structure show immunosuppressive and chemosensitizing effects. Bioorg. Med. Chem. 2017, 25, 5849–5858. [Google Scholar] [CrossRef]
- Sociali, G.; Galeno, L.; Parenti, M.D.; Grozio, A.; Bauer, I.; Passalacqua, M.; Boero, S.; Donadini, A.; Millo, E.; Bellotti, M.; et al. Quinazolinedione SIRT6 inhibitors sensitize cancer cells to chemotherapeutics. Eur. J. Med. Chem. 2015, 102, 530–539. [Google Scholar] [CrossRef]
- He, B.; Hu, J.; Zhang, X.; Lin, H. Thiomyristoyl peptides as cell-permeable Sirt6 inhibitors. Org. Biomol. Chem. 2014, 12, 7498–7502. [Google Scholar] [CrossRef] [Green Version]
- Parenti, M.D.; Grozio, A.; Bauer, I.; Galeno, L.; Damonte, P.; Millo, E.; Sociali, G.; Franceschi, C.; Ballestrero, A.; Bruzzone, S.; et al. Discovery of novel and selective SIRT6 inhibitors. J. Med. Chem. 2014, 57, 4796–4804. [Google Scholar] [CrossRef] [PubMed]








| Substrate Protein Names | Specific Residue(s) | References |
|---|---|---|
| Deacetylase Substrates | ||
| ACSL5, acyl-CoA synthetase long-chain family member 5 | K98, K361, K367 | [16] |
| BECN1, beclin 1 | Not determined | [17] |
| CAV1, caveolin 1 | Not determined | [18] |
| CDH1, cadherin 1 | K135 | [19] |
| CHREBP/MLXIPL, carbohydrate response element binding protein | K672 | [20] |
| CTNNB1, catenin beta 1 | Not determined | [21] |
| DDB2, damage-specific DNA binding protein 2 | K35, K77 | [22] |
| DNMT1, DNA methyltransferase 1 | Not determined | [23] |
| ELF5, E74-like ETS transcription factor 5 | Not determined | [24] |
| ERα/ESR1, estrogen receptor 1 | K171, K299 | [25] |
| ERRγ/ESRRG, estrogen-related receptor gamma | K195 | [26] |
| EZH2, enhancer of Zeste 2 polycomb repressive complex 2 subunit | Not determined | [27] |
| FZD4, frizzled class receptor 4 | Not determined | [28] |
| FOXO1, forkhead box O1 | Not determined | [29,30,31,32,33,34,35,36] |
| FOXO3, forkhead box O3 | K242, K245 | [31,37] |
| FXR/NR1H4, farnesoid X receptor | Not determined | [38] |
| GATA3, GATA binding protein 3 | Not determined | [39] |
| GCN5/KAT2A, lysine acetyltransferase 2A | K549 | [14] |
| HIF1A, hypoxia inducible factor 1A | Not determined | [40] |
| Histone H3 | K9, K18, K56 | [6,7,8,41] |
| HMGB1, high-mobility group box 1 | Not determined | [42] |
| HNRNPA1, heterogenous nuclear ribonucleoprotein A1 | K3, K52, K87, K350 | [43] |
| Ku70/XRCC6, X-ray repair cross complementing 6 | Not determined | [44,45] |
| LXR/NR1H3, liver X receptor | K432 | [20] |
| MRTFA, myocardin-related transcription factor A | Not determined | [46] |
| NAMPT, nicotinamide phosphoribosyltransferase | Not determined | [47] |
| NCOA2, nuclear receptor coactivator 2 | K780 | [48] |
| NFATC4, nuclear factor of activated T cells 4 | Not determined | [49] |
| NF-κB/RELA, NF-kappa-B transcription factor | K310 | [50,51] |
| NOS3, nitric oxide synthase 3 | K494, K497, K504 | [52] |
| NRF2/NFE2L2, NFE2-like BZIP transcription factor 2 | Not determined | [53] |
| P53/TP53, tumor protein P53 | K381 | [54] |
| PER2, period circadian regulator 2 | Not determined | [55] |
| PKM2, pyruvate kinase M2 | K433 | [56] |
| RBBP8, RB binding protein 8 | Not determined | [57] |
| RUNX2, RUNX family transcription factor 2 | Not determined | [58] |
| SMAD2, SMAD family member 2 | K54 | [13] |
| SMAD3, SMAD family member 3 | K333, K378 | [11] |
| SREBP1/SREBF1, sterol regulatory element binding protein 1 | K289 | [20] |
| STAT3, signal transducer and activator of transcription 3 | K685 | [59] |
| STAT5, signal transducer and activator of transcription 5 | K163 | [60] |
| TAZ/WWTR1, WW domain-containing transcription regulator 1 | K39, K54 | [12] |
| TAU/MAPT, microtubule-associated protein Tau | K174 | [61] |
| TRF2, telomeric repeat binding factor 2 | K176, K179, K190 | [62] |
| XBP1, X-box binding protein 1 | K257, K297 | [15] |
| YAP1, Yes1-associated transcriptional regulator | K76, K90, K97, K102, K440 | [12] |
| Deacylase substrates | ||
| RRAS2, RAS-related 2 | K192, K194, K196, K197 | [10] |
| TNFα, tumor necrosis factor alpha | K19, K20 | [9] |
| Mono-ADP ribosyltransferase substrates | ||
| BAF170/SMARCC2, BRG1-associated factor 170 | K312 | [63] |
| KAP1/TRIM28, tripartite motif-containing 28 | Not determined | [64] |
| KDM2A, lysine demethylase 2A | R1020 | [65] |
| LMNA, lamin A/C | Not determined | [66] |
| PARP1, poly(ADP-ribose) polymerase 1 | K521 | [67] |
| SIRT6, sirtuin 6 | Not determined | [5] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, X.C. Sirtuin 6—A Key Regulator of Hepatic Lipid Metabolism and Liver Health. Cells 2023, 12, 663. https://doi.org/10.3390/cells12040663
Dong XC. Sirtuin 6—A Key Regulator of Hepatic Lipid Metabolism and Liver Health. Cells. 2023; 12(4):663. https://doi.org/10.3390/cells12040663
Chicago/Turabian StyleDong, X. Charlie. 2023. "Sirtuin 6—A Key Regulator of Hepatic Lipid Metabolism and Liver Health" Cells 12, no. 4: 663. https://doi.org/10.3390/cells12040663