Arf-like Protein 2 (ARL2) Controls Microtubule Neogenesis during Early Postnatal Photoreceptor Development
Abstract
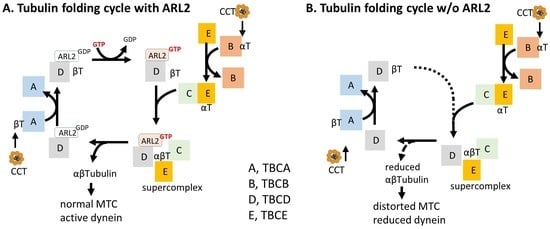
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Generation of Retina- and Rod-Specific Knockout Mice
2.3. Genotyping
2.4. Western Blot
2.5. Confocal Immunohistochemistry
2.6. Electroretinography
2.7. Statistical Analysis
3. Results
3.1. Generation of the Retina-(retArl2−/−) and Rod-Specific (rodArl2−/−) ARL2 Knockouts
3.2. retARL2−/− −/− Outer Nuclear Layers Display Abnormal Histogenesis
3.3. Cytoplasmic Dynein Is Unstable in retArl2−/− Photoreceptors
3.4. retArl2−/− versus rodArl2−/− Electroretinography
3.5. Effect of ARL2 Deletion on Pericentriolar Material
3.6. Microtubules Are in Disarray in the ARL2 Knockout
3.7. Microtubule Cytoskeleton Is Unstable in Both retARL2−/− and retDync1h1−/− Retina
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tamkun, J.W.; Kahn, R.A.; Kissinger, M.; Brizuela, B.J.; Rulka, C.; Scott, M.P.; Kennison, J.A. The arflike gene encodes an essential GTP-binding protein in Drosophila. Proc. Natl. Acad. Sci. USA 1991, 88, 3120–3124. [Google Scholar] [CrossRef] [Green Version]
- Cavenagh, M.M.; Breiner, M.; Schurmann, A.; Rosenwald, A.G.; Terui, T.; Zhang, C.; Randazzo, P.A.; Adams, M.; Joost, H.G.; Kahn, R.A. ADP-ribosylation factor (ARF)-like 3, a new member of the ARF family of GTP-binding proteins cloned from human and rat tissues. J. Biol. Chem. 1994, 269, 18937–18942. [Google Scholar] [CrossRef] [PubMed]
- Schurmann, A.; Breiner, M.; Becker, W.; Huppertz, C.; Kainulainen, H.; Kentrup, H.; Joost, H.G. Cloning of two novel ADP-ribosylation factor-like proteins and characterization of their differential expression in 3T3-L1 cells. J. Biol. Chem. 1994, 269, 15683–15688. [Google Scholar] [CrossRef] [PubMed]
- Sztul, E.; Chen, P.W.; Casanova, J.E.; Cherfils, J.; Dacks, J.B.; Lambright, D.G.; Lee, F.S.; Randazzo, P.A.; Santy, L.C.; Schurmann, A.; et al. ARF GTPases and their GEFs and GAPs: Concepts and challenges. Mol. Biol. Cell 2019, 30, 1249–1271. [Google Scholar] [CrossRef]
- Fisher, S.; Kuna, D.; Caspary, T.; Kahn, R.A.; Sztul, E. ARF family GTPases with links to cilia. Am. J. Physiol. Cell Physiol. 2020, 319, C404–C418. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, M.A.; Stearns, T.; Botstein, D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol. Cell. Biol. 1990, 10, 223–234. [Google Scholar]
- McElver, J.; Patton, D.; Rumbaugh, M.; Liu, C.; Yang, L.J.; Meinke, D. The TITAN5 gene of Arabidopsis encodes a protein related to the ADP ribosylation factor family of GTP binding proteins. Plant Cell 2000, 12, 1379–1392. [Google Scholar] [CrossRef] [Green Version]
- Antoshechkin, I.; Han, M. The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev. Cell 2002, 2, 579–591. [Google Scholar] [CrossRef]
- Li, Y.; Kelly, W.G.; Logsdon, J.M., Jr.; Schurko, A.M.; Harfe, B.D.; Hill-Harfe, K.L.; Kahn, R.A. Functional genomic analysis of the ADP-ribosylation factor family of GTPases: Phylogeny among diverse eukaryotes and function in C. elegans. FASEB J. 2004, 18, 1834–1850. [Google Scholar] [CrossRef]
- Hanzal-Bayer, M.; Renault, L.; Roversi, P.; Wittinghofer, A.; Hillig, R.C. The complex of Arl2-GTP and PDE delta: From structure to function. EMBO J. 2002, 21, 2095–2106. [Google Scholar] [CrossRef] [Green Version]
- Bos, J.L.; Rehmann, H.; Wittinghofer, A. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 2007, 129, 865–877. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.A.; Chen, Y.X.; Miertzschke, M.; Vetter, I.R.; Koerner, C.; Wittinghofer, A. Structural basis for Arl3-specific release of myristoylated ciliary cargo from UNC119. EMBO J. 2012, 31, 4085–4094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watzlich, D.; Vetter, I.; Gotthardt, K.; Miertzschke, M.; Chen, Y.X.; Wittinghofer, A.; Ismail, S. The interplay between RPGR, PDEdelta and Arl2/3 regulate the ciliary targeting of farnesylated cargo. EMBO Rep. 2013, 14, 465–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turn, R.E.; Linnert, J.; Gigante, E.D.; Wolfrum, U.; Caspary, T.; Kahn, R.A. Roles for ELMOD2 and Rootletin in Ciliogenesis. Mol. Biol. Cell 2021, 32, 800–882. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.E.; Schwarz, N.; Zelinger, L.; Stern-Schneider, G.; Shoemark, A.; Spitzbarth, B.; Gross, M.; Laxer, U.; Sosna, J.; Sergouniotis, P.I.; et al. Mutations in ARL2BP, encoding ADP-ribosylation-factor-like 2 binding protein, cause autosomal-recessive retinitis pigmentosa. Am. J. Hum. Genet. 2013, 93, 321–329. [Google Scholar] [CrossRef] [Green Version]
- Moye, A.R.; Bedoni, N.; Cunningham, J.G.; Sanzhaeva, U.; Tucker, E.S.; Mathers, P.; Peter, V.G.; Quinodoz, M.; Paris, L.P.; Coutinho-Santos, L.; et al. Mutations in ARL2BP, a protein required for ciliary microtubule structure, cause syndromic male infertility in humans and mice. PLoS Genet. 2019, 15, e1008315. [Google Scholar] [CrossRef] [Green Version]
- Moye, A.R.; Singh, R.; Kimler, V.A.; Dilan, T.L.; Munezero, D.; Saravanan, T.; Goldberg, A.F.X.; Ramamurthy, V. ARL2BP, a protein linked to retinitis pigmentosa, is needed for normal photoreceptor cilia doublets and outer segment structure. Mol. Biol. Cell 2018, 29, 1590–1598. [Google Scholar] [CrossRef]
- Sharer, J.D.; Kahn, R.A. The ARF-like 2 (ARL2)-binding protein, BART. Purification, cloning, and initial characterization. J. Biol. Chem. 1999, 274, 27553–27561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Humbert, M.C.; Weihbrecht, K.; Searby, C.C.; Li, Y.; Pope, R.M.; Sheffield, V.C.; Seo, S. ARL13B, PDE6D, and CEP164 form a functional network for INPP5E ciliary targeting. Proc. Natl. Acad. Sci. USA 2012, 109, 19691–19696. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.B.; Wu, K.C.; Zhang, X.; Lv, J.N.; Jin, G.H.; Xiang, L.; Chen, J.; Huang, X.F.; Pan, D.; Lu, B.; et al. Whole-exome sequencing identified ARL2 as a novel candidate gene for MRCS (microcornea, rod-cone dystrophy, cataract, and posterior staphyloma) syndrome. Clin. Genet. 2019, 96, 61–71. [Google Scholar] [CrossRef]
- Wright, Z.C.; Loskutov, Y.; Murphy, D.; Stoilov, P.; Pugacheva, E.; Goldberg, A.F.X.; Ramamurthy, V. ADP-Ribosylation Factor-Like 2 (ARL2) regulates cilia stability and development of outer segments in rod photoreceptor neurons. Sci. Rep. 2018, 8, 16967. [Google Scholar] [CrossRef]
- Francis, J.W.; Goswami, D.; Novick, S.J.; Pascal, B.D.; Weikum, E.R.; Ortlund, E.A.; Griffin, P.R.; Kahn, R.A. Nucleotide Binding to ARL2 in the TBCDARL2beta-Tubulin Complex Drives Conformational Changes in beta-Tubulin. J. Mol. Biol. 2017, 429, 3696–3716. [Google Scholar] [CrossRef]
- Fansa, E.K.; Wittinghofer, A. Sorting of lipidated cargo by the Arl2/Arl3 system. Small GTPases 2016, 7, 222–230. [Google Scholar] [CrossRef] [Green Version]
- Schiavon, C.R.; Turn, R.E.; Newman, L.E.; Kahn, R.A. ELMOD2 regulates mitochondrial fusion in a mitofusin-dependent manner, downstream of ARL2. Mol. Biol. Cell 2019, 30, 1198–1213. [Google Scholar] [CrossRef]
- Bowzard, J.B.; Cheng, D.; Peng, J.; Kahn, R.A. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 2007, 282, 17568–17580. [Google Scholar] [CrossRef] [Green Version]
- Turn, R.E.; East, M.P.; Prekeris, R.; Kahn, R.A. The ARF GAP ELMOD2 acts with different GTPases to regulate centrosomal microtubule nucleation and cytokinesis. Mol. Biol. Cell 2020, 31, 2070–2091. [Google Scholar] [CrossRef]
- Newman, L.E.; Schiavon, C.R.; Turn, R.E.; Kahn, R.A. The ARL2 GTPase regulates mitochondrial fusion from the intermembrane space. Cell. Logist. 2017, 7, e1340104. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Feng, Y.; Chen, A.; Li, T.; Huang, S.; Liu, J.; Liu, X.; Liu, Y.; Gao, J.; Yan, D.; et al. Elmod3 knockout leads to progressive hearing loss and abnormalities in cochlear hair cell stereocilia. Hum. Mol. Genet. 2019, 28, 4103–4112. [Google Scholar] [CrossRef]
- Ivanova, A.A.; East, M.P.; Yi, S.L.; Kahn, R.A. Characterization of recombinant ELMOD (cell engulfment and motility domain) proteins as GTPase-activating proteins (GAPs) for ARF family GTPases. J. Biol. Chem. 2014, 289, 11111–11121. [Google Scholar] [CrossRef] [Green Version]
- Tian, G.; Cowan, N.J. Tubulin-specific chaperones: Components of a molecular machine that assembles the alpha/beta heterodimer. Methods Cell Biol. 2013, 115, 155–171. [Google Scholar]
- Tian, G.; Thomas, S.; Cowan, N.J. Effect of TBCD and its regulatory interactor Arl2 on tubulin and microtubule integrity. Cytoskeleton 2010, 67, 706–714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, G.; Huang, Y.; Rommelaere, H.; Vandekerckhove, J.; Ampe, C.; Cowan, N.J. Pathway leading to correctly folded beta-tubulin. Cell 1996, 86, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Cunningham, L.; Marcus, A.I.; Li, Y.; Kahn, R.A. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol. Biol. Cell 2006, 17, 2476–2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, J.W.; Newman, L.E.; Cunningham, L.A.; Kahn, R.A. A Trimer Consisting of the Tubulin-specific Chaperone D (TBCD), Regulatory GTPase ARL2, and beta-Tubulin Is Required for Maintaining the Microtubule Network. J. Biol. Chem. 2017, 292, 4336–4349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhamidipati, A.; Lewis, S.A.; Cowan, N.J. ADP ribosylation factor-like protein 2 (Arl2) regulates the interaction of tubulin-folding cofactor D with native tubulin. J. Cell Biol. 2000, 149, 1087–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuta, Y.; Lagutin, O.; Hogan, B.L.; Oliver, G.C. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. Genesis 2000, 26, 130–132. [Google Scholar] [CrossRef]
- Li, S.; Chen, D.; Sauve, Y.; McCandless, J.; Chen, Y.J.; Chen, C.K. Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis 2005, 41, 73–80. [Google Scholar] [CrossRef]
- Mattapallil, M.J.; Wawrousek, E.F.; Chan, C.C.; Zhao, H.; Roychoudhury, J.; Ferguson, T.A.; Caspi, R.R. The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Investig. Ophthalmol. Vis. Sci. 2012, 53, 2921–2927. [Google Scholar] [CrossRef] [Green Version]
- Reed, M.; Takemaru, K.I.; Ying, G.; Frederick, J.M.; Baehr, W. Deletion of CEP164 in mouse photoreceptors post-ciliogenesis interrupts ciliary intraflagellar transport (IFT). PLoS Genet. 2022, 18, e1010154. [Google Scholar] [CrossRef]
- Dahl, T.M.; Reed, M.; Gerstner, C.D.; Baehr, W. Conditional Deletion of Cytoplasmic Dynein Heavy Chain in Postnatal Photoreceptors. Investig. Ophthalmol. Vis. Sci. 2021, 62, 23. [Google Scholar] [CrossRef]
- Dahl, T.M.; Reed, M.; Gerstner, C.D.; Ying, G.; Baehr, W. Effect of conditional deletion of cytoplasmic dynein heavy chain DYNC1H1 on postnatal photoreceptors. PLoS ONE 2021, 16, e0248354. [Google Scholar] [CrossRef]
- Sharif, A.S.; Gerstner, C.D.; Cady, M.A.; Arshavsky, V.Y.; Mitchell, C.; Ying, G.; Frederick, J.M.; Baehr, W. Deletion of the phosphatase INPP5E in the murine retina impairs photoreceptor axoneme formation and prevents disc morphogenesis. J. Biol. Chem. 2021, 296, 100529. [Google Scholar] [CrossRef]
- Lau, L.; Lee, Y.L.; Sahl, S.J.; Stearns, T.; Moerner, W.E. STED microscopy with optimized labeling density reveals 9-fold arrangement of a centriole protein. Biophys. J. 2012, 102, 2926–2935. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.T.; Chong, W.M.; Wang, W.J.; Mazo, G.; Tanos, B.; Chen, Z.; Tran, T.M.N.; Chen, Y.D.; Weng, R.R.; Huang, C.E.; et al. Super-resolution architecture of mammalian centriole distal appendages reveals distinct blade and matrix functional components. Nat. Commun. 2018, 9, 2023. [Google Scholar] [CrossRef] [Green Version]
- Spektor, A.; Tsang, W.Y.; Khoo, D.; Dynlacht, B.D. Cep97 and CP110 suppress a cilia assembly program. Cell 2007, 130, 678–690. [Google Scholar] [CrossRef] [Green Version]
- Vlijm, R.; Li, X.; Panic, M.; Ruthnick, D.; Hata, S.; Herrmannsdorfer, F.; Kuner, T.; Heilemann, M.; Engelhardt, J.; Hell, S.W.; et al. STED nanoscopy of the centrosome linker reveals a CEP68-organized, periodic rootletin network anchored to a C-Nap1 ring at centrioles. Proc. Natl. Acad. Sci. USA 2018, 115, E2246–E2253. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Li, T. Focus on molecules: Rootletin. Exp. Eye Res. 2006, 83, 1–2. [Google Scholar] [CrossRef]
- Baehr, W.; Hanke-Gogokhia, C.; Sharif, A.; Reed, M.; Dahl, T.M.; Frederick, J.M.; Ying, G. Insights into photoreceptor ciliogenesis revealed by animal models. Prog. Retin. Eye Res. 2019, 71, 26–56. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell. Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef]
- Bosch Grau, M.; Masson, C.; Gadadhar, S.; Rocha, C.; Tort, O.; Marques Sousa, P.; Vacher, S.; Bieche, I.; Janke, C. Alterations in the balance of tubulin glycylation and glutamylation in photoreceptors leads to retinal degeneration. J. Cell Sci. 2017, 130, 938–949. [Google Scholar] [CrossRef] [Green Version]
- Ismail, S.A.; Chen, Y.X.; Rusinova, A.; Chandra, A.; Bierbaum, M.; Gremer, L.; Triola, G.; Waldmann, H.; Bastiaens, P.I.; Wittinghofer, A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat. Chem. Biol. 2011, 7, 942–949. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.A.; Tian, G.; Cowan, N.J. The alpha- and beta-tubulin folding pathways. Trends Cell Biol. 1997, 7, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Nithianantham, S.; Le, S.; Seto, E.; Jia, W.; Leary, J.; Corbett, K.D.; Moore, J.K.; Al-Bassam, J. Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alphabeta-tubulin pool for microtubule dynamics. eLife 2015, 4, e08811. [Google Scholar] [CrossRef]
- Bowzard, J.B.; Sharer, J.D.; Kahn, R.A. Assays used in the analysis of Arl2 and its binding partners. Methods Enzymol. 2005, 404, 453–467. [Google Scholar]
- Cunningham, L.A.; Kahn, R.A. Cofactor D functions as a centrosomal protein and is required for the recruitment of the gamma-tubulin ring complex at centrosomes and organization of the mitotic spindle. J. Biol. Chem. 2008, 283, 7155–7165. [Google Scholar] [CrossRef] [Green Version]
- Yaffe, M.B.; Farr, G.W.; Miklos, D.; Horwich, A.L.; Sternlicht, M.L.; Sternlicht, H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature 1992, 358, 245–248. [Google Scholar] [CrossRef]
- Chen, K.; Koe, C.T.; Xing, Z.B.; Tian, X.; Rossi, F.; Wang, C.; Tang, Q.; Zong, W.; Hong, W.J.; Taneja, R.; et al. Arl2- and Msps-dependent microtubule growth governs asymmetric division. J. Cell Biol. 2016, 212, 661–676. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Jiang, H.; Gu, J.; Tang, Y.; Shen, N.; Jin, Y. MicroRNA-195 targets ADP-ribosylation factor-like protein 2 to induce apoptosis in human embryonic stem cell-derived neural progenitor cells. Cell Death Dis. 2013, 4, e695. [Google Scholar] [CrossRef]
- Ohba, S.; Kamata, K.; Miki-Noumura, T. Stabilization of microtubules by dynein-binding in vitro. Stability of microtubule-dynein complex. Biochim. Biophys. Acta 1993, 1158, 323–332. [Google Scholar] [CrossRef]
- Piontek, K.; Menezes, L.F.; Garcia-Gonzalez, M.A.; Huso, D.L.; Germino, G.G. A critical developmental switch defines the kinetics of kidney cyst formation after loss of Pkd1. Nat. Med. 2007, 13, 1490–1495. [Google Scholar] [CrossRef]
- Dahl, T.M.; Baehr, W. Review: Cytoplasmic dynein motors in photoreceptors. Mol. Vis. 2021, 27, 506–517. [Google Scholar]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gerstner, C.D.; Reed, M.; Dahl, T.M.; Ying, G.; Frederick, J.M.; Baehr, W. Arf-like Protein 2 (ARL2) Controls Microtubule Neogenesis during Early Postnatal Photoreceptor Development. Cells 2023, 12, 147. https://doi.org/10.3390/cells12010147
Gerstner CD, Reed M, Dahl TM, Ying G, Frederick JM, Baehr W. Arf-like Protein 2 (ARL2) Controls Microtubule Neogenesis during Early Postnatal Photoreceptor Development. Cells. 2023; 12(1):147. https://doi.org/10.3390/cells12010147
Chicago/Turabian StyleGerstner, Cecilia D., Michelle Reed, Tiffanie M. Dahl, Guoxin Ying, Jeanne M. Frederick, and Wolfgang Baehr. 2023. "Arf-like Protein 2 (ARL2) Controls Microtubule Neogenesis during Early Postnatal Photoreceptor Development" Cells 12, no. 1: 147. https://doi.org/10.3390/cells12010147