PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli
Abstract
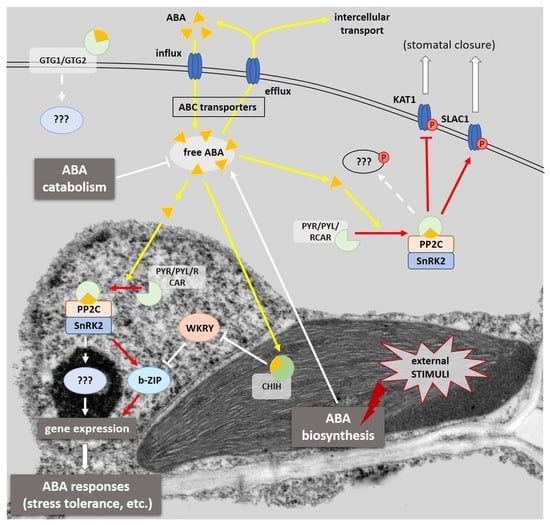
:1. Introduction
2. ABA Signaling Pathway
2.1. The Dependence of the ABA Signaling Pathway on the Expression of ABA Metabolic Genes
2.2. Types of ABA Receptors
2.3. Type 2C Protein Phosphatases—Negative Regulators of ABA Signaling
2.4. Interaction of PYR/PYL/RCAR Receptors with PP2C Phosphatases
2.5. SnRK2 Kinases as Positive Regulators of the ABA Transduction Pathway
3. The Regulation of Gene Expression Dependent on the Presence of ABA
4. Structure, Characteristics, and Functioning of PYR/PYL Receptors
4.1. Structure of the Internal Pocket of PYL Receptors and Their Ability to Bind ABA
4.2. The Degree of Oligomerization of the Structure of PYL Receptors
4.3. Conformational Changes in the PYL Receptor as an Effect of ABA Binding
4.4. PYL Receptors as Essential Elements of the Plant Stress Response: Lessons from the Study of Drought
4.5. Model of PYL Receptor–ABA–PP2C Interaction
4.6. Model of the PP2C–SnRK2 Interaction
4.7. Degradation Pathway for ABA Receptors
5. ABA Signal Termination by Inhibitors
6. SnRK2 Degradation with Inhibitors
Model of HOS15 Participation in the ABA Signaling Pathway
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fidler, J.; Zdunek-Zastocka, E.; Bielawski, W. Regulation of Abscisic Acid Metabolism in Relation to the Dormancy and Germination of Cereal Grains. Acta Soc. Bot. Pol. 2015, 84, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Fidler, J.; Zdunek-Zastocka, E.; Prabucka, B.; Bielawski, W. Abscisic Acid Content and the Expression of Genes Related to Its Metabolism during Maturation of Triticale Grains of Cultivars Differing in Pre-Harvest Sprouting Susceptibility. J. Plant Physiol. 2016, 207, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fidler, J.; Grabowska, A.; Prabucka, B.; Więsyk, A.; Góra-Sochacka, A.; Bielawski, W.; Pojmaj, M.; Zdunek-Zastocka, E. The Varied Ability of Grains to Synthesize and Catabolize ABA Is One of the Factors Affecting Dormancy and Its Release by After-Ripening in Imbibed Triticale Grains of Cultivars with Different Pre-Harvest Sprouting Susceptibilities. J. Plant Physiol. 2018, 226, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Gietler, M.; Fidler, J.; Labudda, M.; Nykiel, M. Abscisic Acid—Enemy or Savior in the Response of Cereals to Abiotic and Biotic Stresses? Int. J. Mol. Sci. 2020, 21, 4607. [Google Scholar] [CrossRef]
- Morkunas, I.; Doğu, M.Z.; Woźniak, A.; Bednarski, W.; Kęsy, J.; Bocianowski, J.; Atar, Ş.H.; Ürün, İ.D.; Labudda, M.; Zydlik, Z.; et al. Profile of Semiquinone Radicals, Phytohormones and Sugars in Pistacia Vera L. Cv. Kirmizi Development. Agronomy 2021, 11, 2115. [Google Scholar] [CrossRef]
- Muszyńska, E.; Tokarz, K.M.; Dziurka, M.; Labudda, M.; Dziurka, K.; Tokarz, B. Photosynthetic Apparatus Efficiency, Phenolic Acid Profiling and Pattern of Chosen Phytohormones in Pseudometallophyte Alyssum Montanum. Sci. Rep. 2021, 11, 4135. [Google Scholar] [CrossRef]
- Formela-Luboińska, M.; Chadzinikolau, T.; Drzewiecka, K.; Jeleń, H.; Bocianowski, J.; Kęsy, J.; Labudda, M.; Jeandet, P.; Morkunas, I. The Role of Sugars in the Regulation of the Level of Endogenous Signaling Molecules during Defense Response of Yellow Lupine to Fusarium Oxysporum. Int. J. Mol. Sci. 2020, 21, 4133. [Google Scholar] [CrossRef]
- Sirko, A.; Wawrzyńska, A.; Brzywczy, J.; Sieńko, M. Control of ABA Signaling and Crosstalk with Other Hormones by the Selective Degradation of Pathway Components. Int. J. Mol. Sci. 2021, 22, 4638. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Koren, O.G. Basic Protein Modules Combining Abscisic Acid and Light Signaling in Arabidopsis. Front. Plant Sci. 2022, 12, 808960. [Google Scholar] [CrossRef]
- Ruiz-Partida, R.; Rosario, S.M.; Lozano-Juste, J. An Update on Crop ABA Receptors. Plants 2021, 10, 1087. [Google Scholar] [CrossRef]
- Kundu, S.; Gantait, S. Abscisic Acid Signal Crosstalk during Abiotic Stress Response. Plant Gene 2017, 11, 61–69. [Google Scholar] [CrossRef]
- Fernando, V.C.D.; Schroeder, D.F. Role of ABA in arabidopsis salt, drought, and desiccation tolerance. In Abiotic and Biotic Stress in Plants—Recent Advances and Future Perspectives; Shanker, A.K., Shanker, C., Eds.; IntechOpen: London, UK, 2016. [Google Scholar] [CrossRef] [Green Version]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanada, K.; Hase, T.; Toyoda, T.; Shinozaki, K.; Okamoto, M. Origin and Evolution of Genes Related to ABA Metabolism and Its Signaling Pathways. J. Plant Res. 2011, 124, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Okamoto, M.; Koshiba, T. ABA biosynthetic and catabolic pathways. In Abscisic Acid: Metabolism, Transport and Signaling; Zhang, D.P., Ed.; Springer: Dordrecht, Germany, 2014; pp. 21–45. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.G.; Pandey, M.; et al. Abscisic Acid Signaling and Abiotic Stress Tolerance in Plants: A Review on Current Knowledge and Future Prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA Perception and Signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.-Q.; Xin, Q.; Cao, Z.; Liu, Z.-Q.; Du, S.-Y.; Mei, C.; Zhao, C.-X.; Wang, X.-F.; Shang, Y.; Jiang, T.; et al. The Magnesium-Chelatase H Subunit Binds Abscisic Acid and Functions in Abscisic Acid Signaling: New Evidence in Arabidopsis. Plant Physiol. 2009, 150, 1940–1954. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.L.; Jiang, L.; Xin, Q.; Liu, Y.; Tan, J.X.; Chen, Z.Z. Structural Basis and Functions of Abscisic Acid Receptors PYLs. Front. Plant Sci. 2015, 6, 88. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.-P.; Wu, Z.-Y.; Li, X.-Y.; Zhao, Z.-X. Purification and Identification of a 42-Kilodalton Abscisic Acid-Specific-Binding Protein from Epidermis of Broad Bean Leaves. Plant Physiol. 2002, 128, 714–725. [Google Scholar] [CrossRef]
- Shen, Y.-Y.; Wang, X.-F.; Wu, F.-Q.; Du, S.-Y.; Cao, Z.; Shang, Y.; Wang, X.-L.; Peng, C.-C.; Yu, X.-C.; Zhu, S.-Y.; et al. The Mg-Chelatase H Subunit Is an Abscisic Acid Receptor. Nature 2006, 443, 823–826. [Google Scholar] [CrossRef]
- Müller, A.H.; Hansson, M. The Barley Magnesium Chelatase 150-KD Subunit Is Not an Abscisic Acid Receptor. Plant Physiol. 2009, 150, 157–166. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Yan, L.; Liu, Z.-Q.; Cao, Z.; Mei, C.; Xin, Q.; Wu, F.-Q.; Wang, X.-F.; Du, S.-Y.; Jiang, T.; et al. The Mg-Chelatase H Subunit of Arabidopsis Antagonizes a Group of WRKY Transcription Repressors to Relieve ABA-Responsive Genes of Inhibition. Plant Cell 2010, 22, 1909–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelstein, R. Abscisic Acid Synthesis and Response. Arabidopsis Book 2013, 11, e0166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, S.; Nelson, D.C.; Assmann, S.M. Two Novel GPCR-Type G Proteins Are Abscisic Acid Receptors in Arabidopsis. Cell 2009, 136, 136–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.-F.; Zhang, D.-P. ABA signal perception and ABA receptors. In Abscisic Acid: Metabolism, Transport and Signaling; Zhang, D.-P., Ed.; Springer: Dordrecht, The Netherlands, 2014; pp. 89–116. ISBN 978-94-017-9424-4. [Google Scholar]
- Alvarez, S.; Roy Choudhury, S.; Hicks, L.M.; Pandey, S. Quantitative Proteomics-Based Analysis Supports a Significant Role of GTG Proteins in Regulation of ABA Response in Arabidopsis Roots. J. Proteome Res. 2013, 12, 1487–1501. [Google Scholar] [CrossRef]
- Jaffé, F.W.; Freschet, G.-E.C.; Valdes, B.M.; Runions, J.; Terry, M.J.; Williams, L.E. G Protein-Coupled Receptor-Type G Proteins Are Required for Light-Dependent Seedling Growth and Fertility in Arabidopsis. Plant Cell 2012, 24, 3649–3668. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.-F.F.; et al. Abscisic Acid Inhibits Type 2C Protein Phosphatases via the PYR/PYL Family of START Proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C Phosphatase Activity Function as Abscisic Acid Sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Tian, X.; Wang, Z.; Li, X.; Lv, T.; Liu, H.; Wang, L.; Niu, H.; Bu, Q. Characterization and Functional Analysis of Pyrabactin Resistance-Like Abscisic Acid Receptor Family in Rice. Rice 2015, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, F.; Kuromori, T.; Sato, H.; Shinozaki, K. Regulatory Gene Networks in Drought Stress Responses and Resistance in Plants. Adv. Exp. Med. Biol. 2018, 1081, 189–214. [Google Scholar] [CrossRef]
- Santiago, J.; Dupeux, F.; Round, A.; Antoni, R.; Park, S.-Y.; Jamin, M.; Cutler, S.R.; Rodriguez, P.L.; Márquez, J.A. The Abscisic Acid Receptor PYR1 in Complex with Abscisic Acid. Nature 2009, 462, 665–668. [Google Scholar] [CrossRef]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.-Y.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.-K. In Vitro Reconstitution of an Abscisic Acid Signalling Pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, K.; Yamaguchi-Shinozaki, K. ABA Signaling in Stress-Response and Seed Development. Plant Cell Rep. 2013, 32, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Rodrigues, A.; Pizzio, G.A.; Rodriguez, P.L. Selective Inhibition of Clade A Phosphatases Type 2C by PYR/PYL/RCAR Abscisic Acid Receptors. Plant Physiol. 2012, 158, 970–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaskara, G.B.; Nguyen, T.T.; Verslues, P.E. Unique Drought Resistance Functions of the Highly ABA-Induced Clade A Protein Phosphatase 2Cs. Plant Physiol. 2012, 160, 379–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, J.; Merlot, S.; Giraudat, J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 Genes Encode Homologous Protein Phosphatases 2C Involved in Abscisic Acid Signal Transduction. Plant Cell 1997, 9, 759–771. [Google Scholar] [CrossRef] [Green Version]
- Saez, A.; Apostolova, N.; Gonzalez-Guzman, M.; Gonzalez-Garcia, M.P.; Nicolas, C.; Lorenzo, O.; Rodriguez, P.L. Gain-of-Function and Loss-of-Function Phenotypes of the Protein Phosphatase 2C HAB1 Reveal Its Role as a Negative Regulator of Abscisic Acid Signalling. Plant J. 2004, 37, 354–369. [Google Scholar] [CrossRef]
- Kashtoh, H.; Baek, K.-H. Structural and Functional Insights into the Role of Guard Cell Ion Channels in Abiotic Stress-Induced Stomatal Closure. Plants 2021, 10, 2774. [Google Scholar] [CrossRef]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. ABA Control of Plant Macroelement Membrane Transport Systems in Response to Water Deficit and High Salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef]
- Merlot, S.; Mustilli, A.-C.; Genty, B.; North, H.; Lefebvre, V.; Sotta, B.; Vavasseur, A.; Giraudat, J. Use of Infrared Thermal Imaging to Isolate Arabidopsis Mutants Defective in Stomatal Regulation. Plant J. 2002, 30, 601–609. [Google Scholar] [CrossRef] [Green Version]
- Brandt, B.; Brodsky, D.E.; Xue, S.; Negi, J.; Iba, K.; Kangasjärvi, J.; Ghassemian, M.; Stephan, A.B.; Hu, H.; Schroeder, J.I. Reconstitution of Abscisic Acid Activation of SLAC1 Anion Channel by CPK6 and OST1 Kinases and Branched ABI1 PP2C Phosphatase Action. Proc. Natl. Acad. Sci. USA 2012, 109, 10593–10598. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Lan, W.; Buchanan, B.B.; Luan, S. A Protein Kinase-Phosphatase Pair Interacts with an Ion Channel to Regulate ABA Signaling in Plant Guard Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 21419–21424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 Protein Phosphatases 2C Act in a Negative Feedback Regulatory Loop of the Abscisic Acid Signalling Pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Lan, W.-Z.; Lee, S.-C.; Che, Y.-F.; Jiang, Y.-Q.; Luan, S. Mechanistic Analysis of AKT1 Regulation by the CBL–CIPK–PP2CA Interactions. Mol. Plant 2011, 4, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Ross, K.E.; Kaldis, P.; Solomon, M.J. Dephosphorylation of Cyclin-Dependent Kinases by Type 2C Protein Phosphatases. Genes Dev. 1999, 13, 2946–2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-G.; Fu, F.-L.; Yu, H.-Q.; Hu, T.; Zhang, Y.-Y.; Tao, Y.; Zhu, J.-K.; Zhao, Y.; Li, W.-C. Interaction Network of Core ABA Signaling Components in Maize. Plant Mol. Biol. 2018, 96, 245–263. [Google Scholar] [CrossRef] [PubMed]
- Tischer, S.V.; Wunschel, C.; Papacek, M.; Kleigrewe, K.; Hofmann, T.; Christmann, A.; Grill, E. Combinatorial Interaction Network of Abscisic Acid Receptors and Coreceptors from Arabidopsis Thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 10280–10285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, S.; Min, M.K.; Lee, S.-Y.; Lim, C.W.; Bhatnagar, N.; Lee, Y.; Shin, D.; Chung, K.Y.; Lee, S.C.; Kim, B.-G.; et al. Modulation of ABA Signaling by Altering VxGΦL Motif of PP2Cs in Oryza Sativa. Mol. Plant 2017, 10, 1190–1205. [Google Scholar] [CrossRef] [Green Version]
- Saruhashi, M.; Kumar Ghosh, T.; Arai, K.; Ishizaki, Y.; Hagiwara, K.; Komatsu, K.; Shiwa, Y.; Izumikawa, K.; Yoshikawa, H.; Umezawa, T.; et al. Plant Raf-like Kinase Integrates Abscisic Acid and Hyperosmotic Stress Signaling Upstream of SNF1-Related Protein Kinase2. Proc. Natl. Acad. Sci. USA 2015, 112, E6388–E6396. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Chen, S.; Harmon, A.C. Protein Phosphorylation in Stomatal Movement. Plant Signal. Behav. 2014, 9, e972845. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wang, X.-Q.; Watson, M.B.; Assmann, S.M. Regulation of Abscisic Acid-Induced Stomatal Closure and Anion Channels by Guard Cell AAPK Kinase. Science 2000, 287, 300–303. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, R.; Umezawa, T.; Mizoguchi, T.; Takahashi, S.; Takahashi, F.; Shinozaki, K. The Regulatory Domain of SRK2E/OST1/SnRK2.6 Interacts with ABI1 and Integrates Abscisic Acid (ABA) and Osmotic Stress Signals Controlling Stomatal Closure in Arabidopsis. J. Biol. Chem. 2006, 281, 5310–5318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 Protein Kinases Are the Main Positive Regulators of Abscisic Acid Signaling in Response to Water Stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 Protein Kinases—Key Regulators of Plant Response to Abiotic Stresses. OMICS 2011, 15, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Ren, R.; Zhang, Y.; Pang, Y.; Yan, C.; Gong, X.; He, Y.; Li, W.; Miao, D.; Hao, Q.; et al. Molecular Mechanism for Inhibition of a Critical Component in the Arabidopsis Thaliana Abscisic Acid Signal Transduction Pathways, SnRK2.6, by Protein Phosphatase ABI1. J. Biol. Chem. 2012, 287, 794–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-Dependent and ABA-Independent Signaling in Response to Osmotic Stress in Plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Kang, J.; Choi, H.; Im, M.; Kim, S.Y. Arabidopsis Basic Leucine Zipper Proteins That Mediate Stress-Responsive Abscisic Acid Signaling. Plant Cell 2002, 14, 343–357. [Google Scholar] [CrossRef]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal Role of the AREB/ABF-SnRK2 Pathway in ABRE-Mediated Transcription in Response to Osmotic Stress in Plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Sayama, H.; Kidokoro, S.; Maruyama, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1, AREB2, and ABF3 Are Master Transcription Factors That Cooperatively Regulate ABRE-Dependent ABA Signaling Involved in Drought Stress Tolerance and Require ABA for Full Activation. Plant J. 2010, 61, 672–685. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF Transcription Factors Function Predominantly in Gene Expression Downstream of SnRK2 Kinases in Abscisic Acid Signalling in Response to Osmotic Stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Fujita, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. ABA-Mediated Transcriptional Regulation in Response to Osmotic Stress in Plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.; Li, Y.; Zheng, N.; Chen, H.; Zhao, Q.; Gao, T.; Guo, H.; Xie, Q. SDIR1 Is a RING Finger E3 Ligase That Positively Regulates Stress-Responsive Abscisic Acid Signaling in Arabidopsis. Plant Cell 2007, 19, 1912–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radauer, C.; Lackner, P.; Breiteneder, H. The Bet v 1 Fold: An Ancient, Versatile Scaffold for Binding of Large, Hydrophobic Ligands. BMC Evol. Biol. 2008, 8, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klingler, J.P.; Batelli, G.; Zhu, J.-K. ABA Receptors: The START of a New Paradigm in Phytohormone Signalling. J. Exp. Bot. 2010, 61, 3199–3210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melcher, K.; Ng, L.-M.; Zhou, X.E.; Soon, F.-F.; Xu, Y.; Suino-Powell, K.M.; Park, S.-Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A Gate–Latch–Lock Mechanism for Hormone Signalling by Abscisic Acid Receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyazono, K.-I.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.-J.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y.; et al. Structural Basis of Abscisic Acid Signalling. Nature 2009, 462, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, N.; Hitomi, K.; Arvai, A.S.; Rambo, R.P.; Hitomi, C.; Cutler, S.R.; Schroeder, J.I.; Getzoff, E.D. Structural Mechanism of Abscisic Acid Binding and Signaling by Dimeric PYR1. Science 2009, 326, 1373–1379. [Google Scholar] [CrossRef] [Green Version]
- Yin, P.; Fan, H.; Hao, Q.; Yuan, X.; Wu, D.; Pang, Y.; Yan, C.; Li, W.; Wang, J.; Yan, N. Structural Insights into the Mechanism of Abscisic Acid Signaling by PYL Proteins. Nat. Struct. Mol. Biol. 2009, 16, 1230–1236. [Google Scholar] [CrossRef]
- Rodriguez, P.L.; Lozano-Juste, J.; Albert, A. PYR/PYL/RCAR ABA receptors. In Advances in Botanical Research, Abscisc Acid in Plants; Seo, M., Marion-Poll, A., Eds.; Academic Press: London, UK, 2019; Volume 92, pp. 51–82. [Google Scholar] [CrossRef]
- Dalal, M.; Chinnusamy, V. ABA receptors: Prospects for enhancing biotic and abiotic stress tolerance of crops. In Elucidation of Abiotic Stress Signaling in Plants: Functional Genomics Perspectives, Volume 1; Pandey, G.K., Ed.; Springer: New York, NY, USA, 2015; pp. 271–298. ISBN 978-1-4939-2211-6. [Google Scholar]
- Santiago, J.; Dupeux, F.; Betz, K.; Antoni, R.; Gonzalez-Guzman, M.; Rodriguez, L.; Márquez, J.A.; Rodriguez, P.L. Structural Insights into PYR/PYL/RCAR ABA Receptors and PP2Cs. Plant Sci. 2012, 182, 3–11. [Google Scholar] [CrossRef] [Green Version]
- Hao, Q.; Yin, P.; Li, W.; Wang, L.; Yan, C.; Lin, Z.; Wu, J.Z.; Wang, J.; Yan, S.F.; Yan, N. The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins. Mol. Cell 2011, 42, 662–672. [Google Scholar] [CrossRef]
- He, Y.; Hao, Q.; Li, W.; Yan, C.; Yan, N.; Yin, P. Identification and Characterization of ABA Receptors in Oryza Sativa. PLoS ONE 2014, 9, e95246. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chan, Z.; Xing, L.; Liu, X.; Hou, Y.-J.; Chinnusamy, V.; Wang, P.; Duan, C.; Zhu, J.-K. The Unique Mode of Action of a Divergent Member of the ABA-Receptor Protein Family in ABA and Stress Signaling. Cell Res. 2013, 23, 1380–1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyakawa, T.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Tanokura, M. Structure and Function of Abscisic Acid Receptors. Trends Plant Sci. 2013, 18, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Frackenpohl, J.; Decker, L.J.B.; Dittgen, J.; Freigang, J.; Génix, P.; Helmke, H.; Lange, G.; Luemmen, P.; Schmidt, J.; Schmutzler, D.; et al. Tetrahydroquinolinyl Phosphinamidates and Phosphonamidates Enhancing Tolerance towards Drought Stress in Crops via Interaction with ABA Receptor Proteins. Bioorg. Med. Chem. 2020, 28, 115725. [Google Scholar] [CrossRef] [PubMed]
- Kido, É.A.; Ferreira-Neto, J.R.; da Silva, M.D.; Santos, V.E.P.; da Filho, J.L.B.S.; Benko-Iseppon, A. Osmoprotectant-related genes in plants under abiotic stress: Expression dynamics, in silico genome mapping, and biotechnology. In Osmoprotectant-Mediated Abiotic Stress Tolerance in Plants; Hossain, M.A., Kumar, V., Burritt, D.J., Fujita, M., Mäkelä, P.S.A., Eds.; Springer International Publishing: Cham, Germany, 2019; pp. 1–40. [Google Scholar] [CrossRef]
- Labudda, M.; Azam, F.M.S. Glutathione-Dependent Responses of Plants to Drought: A Review. Acta Soc. Bot. Pol. 2014, 83, 3–12. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Zhong, J.; Sun, X.; Wang, B.; Terzaghi, W.; Dai, M. The Maize ABA Receptors ZmPYL8, 9, and 12 Facilitate Plant Drought Resistance. Front. Plant Sci. 2018, 9, 422. [Google Scholar] [CrossRef]
- Gupta, M.K.; Lenka, S.K.; Gupta, S.; Rawal, R.K. Agonist, Antagonist and Signaling Modulators of ABA Receptor for Agronomic and Post-Harvest Management. Plant Physiol. Biochem. 2020, 148, 10–25. [Google Scholar] [CrossRef]
- Zhang, Q.; Kong, X.; Yu, Q.; Ding, Y.; Li, X.; Yang, Y. Responses of PYR/PYL/RCAR ABA Receptors to Contrasting Stresses, Heat and Cold in Arabidopsis. Plant Signal. Behav. 2019, 14, 1670596. [Google Scholar] [CrossRef]
- Fujii, H. Abscisic acid implication in plant growth and stress responses. In Phytohormones: A Window to Metabolism, Signaling and Biotechnological Applications; Tran, L.-S.P., Pal, S., Eds.; Springer: New York, NY, USA, 2014; pp. 37–54. ISBN 978-1-4939-0491-4. [Google Scholar]
- Fan, W.; Zhao, M.; Li, S.; Bai, X.; Li, J.; Meng, H.; Mu, Z. Contrasting Transcriptional Responses of PYR1/PYL/RCAR ABA Receptors to ABA or Dehydration Stress between Maize Seedling Leaves and Roots. BMC Plant Biol. 2016, 16, 99. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA Receptor PYL9 Promotes Drought Resistance and Leaf Senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [Green Version]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Liu, Y.; Li, R. Precise Editing of the OsPYL9 Gene by RNA-Guided Cas9 Nuclease Confers Enhanced Drought Tolerance and Grain Yield in Rice (Oryza Sativa L.) by Regulating Circadian Rhythm and Abiotic Stress Responsive Proteins. Int. J. Mol. Sci. 2020, 21, 7854. [Google Scholar] [CrossRef]
- Lenka, S.K.; Muthusamy, S.K.; Chinnusamy, V.; Bansal, K.C. Ectopic Expression of Rice PYL3 Enhances Cold and Drought Tolerance in Arabidopsis Thaliana. Mol. Biotechnol. 2018, 60, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Dalal, M.; Inupakutika, M. Transcriptional Regulation of ABA Core Signaling Component Genes in Sorghum (Sorghum Bicolor L. Moench). Mol. Breeding 2014, 34, 1517–1525. [Google Scholar] [CrossRef]
- Bai, G.; Xie, H.; Yao, H.; Li, F.; Chen, X.; Zhang, Y.; Xiao, B.; Yang, J.; Li, Y.; Yang, D.-H. Genome-Wide Identification and Characterization of ABA Receptor PYL/RCAR Gene Family Reveals Evolution and Roles in Drought Stress in Nicotiana Tabacum. BMC Genom. 2019, 20, 575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boneh, U.; Biton, I.; Zheng, C.; Schwartz, A.; Ben-Ari, G. Characterization of Potential ABA Receptors in Vitis Vinifera. Plant Cell Rep. 2012, 31, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Jang, G.; Um, T.; Kim, J.-K.; Lee, J.S.; Do Choi, Y. The Soluble ABA Receptor PYL8 Regulates Drought Resistance by Controlling ABA Signaling in Arabidopsis. Plant Biotechnol. Rep. 2015, 9, 319–330. [Google Scholar] [CrossRef]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR Receptors Play a Major Role in Quantitative Regulation of Stomatal Aperture and Transcriptional Response to Abscisic Acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [Green Version]
- Dupeux, F.; Antoni, R.; Betz, K.; Santiago, J.; Gonzalez-Guzman, M.; Rodriguez, L.; Rubio, S.; Park, S.-Y.; Cutler, S.R.; Rodriguez, P.L.; et al. Modulation of Abscisic Acid Signaling in Vivo by an Engineered Receptor-Insensitive Protein Phosphatase Type 2C Allele. Plant Physiol. 2011, 156, 106–116. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, J.; Mimura, N.; Okamoto, M.; Yajima, S.; Sue, M.; Akiyama, T.; Monda, K.; Iba, K.; Ohnishi, T.; Todoroki, Y. Structure-Based Chemical Design of Abscisic Acid Antagonists That Block PYL-PP2C Receptor Interactions. ACS Chem. Biol. 2018, 13, 1313–1321. [Google Scholar] [CrossRef]
- Joshi-Saha, A.; Valon, C.; Leung, J. A Brand New START: Abscisic Acid Perception and Transduction in the Guard Cell. Sci. Signal. 2011, 4, re4. [Google Scholar] [CrossRef]
- Leung, J.; Bouvier-Durand, M.; Morris, P.-C.; Guerrier, D.; Chefdor, F.; Giraudat, J. Arabidopsis ABA Response Gene ABI1: Features of a Calcium-Modulated Protein Phosphatase. Science 1994, 264, 1448–1452. [Google Scholar] [CrossRef]
- Gosti, F.; Beaudoin, N.; Serizet, C.; Webb, A.A.R.; Vartanian, N.; Giraudat, J. ABI1 Protein Phosphatase 2C Is a Negative Regulator of Abscisic Acid Signaling. Plant Cell 1999, 11, 1897–1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soon, F.-F.; Ng, L.-M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.H.E.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J.; et al. Molecular Mimicry Regulates ABA Signaling by SnRK2 Kinases and PP2C Phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belin, C.; de Franco, P.-O.; Bourbousse, C.; Chaignepain, S.; Schmitter, J.-M.; Vavasseur, A.; Giraudat, J.; Barbier-Brygoo, H.; Thomine, S. Identification of Features Regulating OST1 Kinase Activity and OST1 Function in Guard Cells. Plant Physiol. 2006, 141, 1316–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Righetto, G.L.; Sriranganadane, D.; Halabelian, L.; Chiodi, C.G.; Elkins, J.M.; Massirer, K.B.; Gileadi, O.; Menossi, M.; Couñago, R.M. The C-Terminal Domains SnRK2 Box and ABA Box Have a Role in Sugarcane SnRK2s Auto-Activation and Activity. Front. Plant Sci. 2019, 10, 1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic Acid Perception and Signaling: Structural Mechanisms and Applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Pardo, J.M.; Yun, D.-J. Desensitization of ABA-Signaling: The Swing From Activation to Degradation. Front. Plant Sci. 2020, 11, 379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-León, M.; Cuyas, L.; El-Moneim, D.A.; Rodriguez, L.; Belda-Palazón, B.; Sanchez-Quant, E.; Fernández, Y.; Roux, B.; Zamarreño, Á.M.; García-Mina, J.M.; et al. Arabidopsis ALIX Regulates Stomatal Aperture and Turnover of Abscisic Acid Receptors. Plant Cell 2019, 31, 2411–2429. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Lou, L.; Tian, M.; Li, Q.; Ding, Y.; Cao, X.; Wu, Y.; Belda-Palazon, B.; Rodriguez, P.L.; Yang, S.; et al. ESCRT-I Component VPS23A Affects ABA Signaling by Recognizing ABA Receptors for Endosomal Degradation. Mol. Plant 2016, 9, 1570–1582. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhao, L.; Zhang, M.; Zafar, S.A.; Fang, J.; Li, M.; Zhang, W.; Li, X. Arabidopsis E3 Ubiquitin Ligases PUB22 and PUB23 Negatively Regulate Drought Tolerance by Targeting ABA Receptor PYL9 for Degradation. Int. J. Mol. Sci. 2017, 18, 1841. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Xie, Q. Ubiquitin-Proteasome System in ABA Signaling: From Perception to Action. Mol. Plant 2016, 9, 21–33. [Google Scholar] [CrossRef] [Green Version]
- Santner, A.; Estelle, M. The Ubiquitin-Proteasome System Regulates Plant Hormone Signaling. Plant J. 2010, 61, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Ludwików, A. Targeting Proteins for Proteasomal Degradation—A New Function of Arabidopsis ABI1 Protein Phosphatase 2C. Front. Plant Sci. 2015, 6, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Lim, C.J.; Shen, M.; Park, H.J.; Cha, J.-Y.; Iniesto, E.; Rubio, V.; Mengiste, T.; Zhu, J.-K.; Bressan, R.A.; et al. Epigenetic Switch from Repressive to Permissive Chromatin in Response to Cold Stress. Proc. Natl. Acad. Sci. USA 2018, 115, E5400–E5409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, A.; Kim, J.K.; Jan, M.; Khan, H.A.; Khan, I.U.; Shen, M.; Park, J.; Lim, C.J.; Hussain, S.; Baek, D.; et al. Rheostatic Control of ABA Signaling through HOS15-Mediated OST1 Degradation. Mol. Plant 2019, 12, 1447–1462. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fidler, J.; Graska, J.; Gietler, M.; Nykiel, M.; Prabucka, B.; Rybarczyk-Płońska, A.; Muszyńska, E.; Morkunas, I.; Labudda, M. PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli. Cells 2022, 11, 1352. https://doi.org/10.3390/cells11081352
Fidler J, Graska J, Gietler M, Nykiel M, Prabucka B, Rybarczyk-Płońska A, Muszyńska E, Morkunas I, Labudda M. PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli. Cells. 2022; 11(8):1352. https://doi.org/10.3390/cells11081352
Chicago/Turabian StyleFidler, Justyna, Jakub Graska, Marta Gietler, Małgorzata Nykiel, Beata Prabucka, Anna Rybarczyk-Płońska, Ewa Muszyńska, Iwona Morkunas, and Mateusz Labudda. 2022. "PYR/PYL/RCAR Receptors Play a Vital Role in the Abscisic-Acid-Dependent Responses of Plants to External or Internal Stimuli" Cells 11, no. 8: 1352. https://doi.org/10.3390/cells11081352