Research Status of the Orphan G Protein Coupled Receptor 158 and Future Perspectives
Abstract
:1. Introduction
2. Discovery of GPR158
3. Structure of GPR158
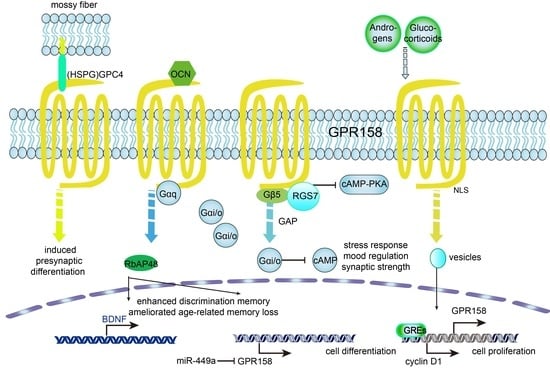
4. Roles of GPR158 in Cancer
- (1)
- Deregulating cellular metabolism: An mRNA microarray study on the subnuclear structures of the mouse brain suggested that habenular GPR158 might be involved in food consumption and energy expenditure (EE) [67]. Single nucleotide polymorphisms (SNPs) of GPR158 were found to be associated with a lower energy expenditure (EE) and adiposity in Native Americans [68]. As a known intracellular interacting protein of GPR158, RGS7 is at an obesity locus in humans [69] and as a putative agonist of GPR158, OCN mediates insulin signals in glucose metabolism [70], which indicated that GPR158 might influence tumor development and neuropsychiatric diseases through energy metabolism.
- (2)
- Avoiding immune destruction: SNPs of the GPR158 gene were shown to be potentially liked to humoral immunity to smallpox vaccination [71] and to hepatitis C virus (HCV) clearance in patients of European and African ancestry [72]. These findings expand the relationship existing between GPR158 and neuronal activity towards its possible role in neuro-immune cross-talk.
- (3)
5. Roles of GPR158 in Affective Disorders
6. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hauser, A.S.; Attwood, M.M.; Rask-Andersen, M.; Schiöth, H.B.; Gloriam, D.E. Trends in GPCR drug discovery: New agents, targets and indications. Nat. Rev. Drug Discov. 2017, 16, 829–842. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A.S.; Chavali, S.; Masuho, I.; Jahn, L.J.; Martemyanov, K.A.; Gloriam, D.E.; Babu, M.M. Pharmacogenomics of GPCR Drug Targets. Cell 2018, 172, 41–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stevens, R.C.; Cherezov, V.; Katritch, V.; Abagyan, R.; Kuhn, P.; Rosen, H.; Wüthrich, K. The GPCR Network: A large-scale collaboration to determine human GPCR structure and function. Nat. Rev. Drug Discov. 2013, 12, 25–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredriksson, R.; Lagerström, M.C.; Lundin, L.G.; Schiöth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 2003, 63, 1256–1272. [Google Scholar] [CrossRef] [Green Version]
- Pin, J.P.; Bettler, B. Organization and functions of mGlu and GABA(B) receptor complexes. Nature 2016, 540, 60–68. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, S.; Xu, F. Structural and Druggability Landscape of Frizzled G Protein-Coupled Receptors. Trends Biochem. Sci. 2018, 43, 1033–1046. [Google Scholar] [CrossRef]
- Bassilana, F.; Nash, M.; Ludwig, M.G. Adhesion G protein-coupled receptors: Opportunities for drug discovery. Nat. Rev. Drug Discov. 2019, 18, 869–884. [Google Scholar] [CrossRef]
- Kooistra, A.J.; Mordalski, S.; Pándy-Szekeres, G.; Esguerra, M.; Mamyrbekov, A.; Munk, C.; Keserű, G.M.; Gloriam, D.E. GPCRdb in 2021: Integrating GPCR sequence, structure and function. Nucleic Acids Res. 2021, 49, D335–D343. [Google Scholar] [CrossRef]
- Pándy-Szekeres, G.; Esguerra, M.; Hauser, A.S.; Caroli, J.; Munk, C.; Pilger, S.; Keserű, G.M.; Kooistra, A.J.; Gloriam, D.E. The G protein database, GproteinDb. Nucleic Acids Res. 2022, 50, D518–D525. [Google Scholar] [CrossRef]
- Insel, P.A.; Wilderman, A.; Zambon, A.C.; Snead, A.N.; Murray, F.; Aroonsakool, N.; McDonald, D.S.; Zhou, S.; McCann, T.; Zhang, L. G Protein-Coupled Receptor (GPCR) Expression in Native Cells: “Novel” endoGPCRs as Physiologic Regulators and Therapeutic Targets. Mol. Pharmacol. 2015, 88, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Mo, X.L.; Yang, Z.; Tao, Y.X. Targeting GPR119 for the potential treatment of type 2 diabetes mellitus. Prog. Mol. Biol. Transl. Sci. 2014, 121, 95–131. [Google Scholar] [PubMed]
- Morgan, R.G.; Mortensson, E.; Williams, A.C. Targeting LGR5 in Colorectal Cancer: Therapeutic gold or too plastic? Br. J. Cancer 2018, 118, 1410–1418. [Google Scholar] [CrossRef] [PubMed]
- Divorty, N.; Mackenzie, A.E.; Nicklin, S.A.; Milligan, G. G protein-coupled receptor 35: An emerging target in inflammatory and cardiovascular disease. Front. Pharmacol. 2015, 6, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planell, N.; Masamunt, M.C.; Leal, R.F.; Rodríguez, L.; Esteller, M.; Lozano, J.J.; Ramírez, A.; Ayrizono, M.L.S.; Coy, C.S.R.; Alfaro, I.; et al. Usefulness of Transcriptional Blood Biomarkers as a Non-invasive Surrogate Marker of Mucosal Healing and Endoscopic Response in Ulcerative Colitis. J. Crohn’s Colitis 2017, 11, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Thomson, P.A.; Wray, N.R.; Thomson, A.M.; Dunbar, D.R.; Grassie, M.A.; Condie, A.; Walker, M.T.; Smith, D.J.; Pulford, D.J.; Muir, W.; et al. Sex-specific association between bipolar affective disorder in women and GPR50, an X-linked orphan G protein-coupled receptor. Mol. Psychiatry 2005, 10, 470–478. [Google Scholar] [CrossRef]
- Patel, N.; Itakura, T.; Jeong, S.; Liao, C.P.; Roy-Burman, P.; Zandi, E.; Groshen, S.; Pinski, J.; Coetzee, G.A.; Gross, M.E.; et al. Expression and functional role of orphan receptor GPR158 in prostate cancer growth and progression. PLoS ONE 2015, 10, e0117758. [Google Scholar] [CrossRef]
- Li, N.; Zhang, Y.; Sidlauskas, K.; Ellis, M.; Evans, I.; Frankel, P.; Lau, J.; El-Hassan, T.; Guglielmi, L.; Broni, J.; et al. Inhibition of GPR158 by microRNA-449a suppresses neural lineage of glioma stem/progenitor cells and correlates with higher glioma grades. Oncogene 2018, 37, 4313–4333. [Google Scholar] [CrossRef] [Green Version]
- Sutton, L.P.; Orlandi, C.; Song, C.; Oh, W.C.; Muntean, B.S.; Xie, K.; Filippini, A.; Xie, X.; Satterfield, R.; Yaeger, J.D.W.; et al. Orphan receptor GPR158 controls stress-induced depression. eLife 2018, 7, e33273. [Google Scholar] [CrossRef]
- Orlandi, C.; Sutton, L.P.; Muntean, B.S.; Song, C.; Martemyanov, K.A. Homeostatic cAMP regulation by the RGS7 complex controls depression-related behaviors. Neuropsychopharmacology 2019, 44, 642–653. [Google Scholar] [CrossRef]
- Bjarnadottir, T.K.; Fredriksson, R.; Schioth, H.B. The gene repertoire and the common evolutionary history of glutamate, pheromone (V2R), taste(1) and other related G protein-coupled receptors. Gene 2005, 362, 70–84. [Google Scholar] [CrossRef]
- Petzold, A.; Reichwald, K.; Groth, M.; Taudien, S.; Hartmann, N.; Priebe, S.; Shagin, D.; Englert, C.; Platzer, M. The transcript catalogue of the short-lived fish Nothobranchius furzeri provides insights into age-dependent changes of mRNA levels. BMC Genom. 2013, 14, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peachey, N.S.; Ray, T.A.; Florijn, R.; Rowe, L.B.; Sjoerdsma, T.; Contreras-Alcantara, S.; Baba, K.; Tosini, G.; Pozdeyev, N.; Iuvone, P.M.; et al. GPR179 is required for depolarizing bipolar cell function and is mutated in autosomal-recessive complete congenital stationary night blindness. Am. J. Hum. Genet. 2012, 90, 331–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, H.A.; Orlandi, C.; Martemyanov, K.A. Beyond the Ligand: Extracellular and Transcellular G Protein-Coupled Receptor Complexes in Physiology and Pharmacology. Pharmacol. Rev. 2019, 71, 503–519. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, C.; Posokhova, E.; Masuho, I.; Ray, T.A.; Hasan, N.; Gregg, R.G.; Martemyanov, K.A. GPR158/179 regulate G protein signaling by controlling localization and activity of the RGS7 complexes. J. Cell Biol. 2012, 197, 711–719. [Google Scholar] [CrossRef] [Green Version]
- Gold, S.J.; Ni, Y.G.; Dohlman, H.G.; Nestler, E.J. Regulators of G-protein signaling (RGS) proteins: Region-specific expression of nine subtypes in rat brain. J. Neurosci. 1997, 17, 8024–8037. [Google Scholar] [CrossRef] [Green Version]
- Hajj, M.; De Vita, T.; Vol, C.; Renassia, C.; Bologna, J.C.; Brabet, I.; Cazade, M.; Pastore, M.; Blahos, J.; Labesse, G.; et al. Nonclassical Ligand-Independent Regulation of Go Protein by an Orphan Class C G-Protein–Coupled Receptor. Mol. Pharmacol. 2019, 96, 233–246. [Google Scholar] [CrossRef]
- Calebiro, D.; Koszegi, Z.; Lanoiselée, Y.; Miljus, T.; O'Brien, S. G protein-coupled receptor-G protein interactions: A single-molecule perspective. Physiol. Rev. 2021, 101, 857–906. [Google Scholar] [CrossRef]
- Patel, N.; Itakura, T.; Gonzalez, J.M., Jr.; Schwartz, S.G.; Fini, M.E. GPR158, an orphan member of G protein-coupled receptor Family C: Glucocorticoid-stimulated expression and novel nuclear role. PLoS ONE 2013, 8, e57843. [Google Scholar] [CrossRef]
- Condomitti, G.; Wierda, K.D.; Schroeder, A.; Rubio, S.E.; Vennekens, K.M.; Orlandi, C.; Martemyanov, K.A.; Gounko, N.V.; Savas, J.N.; de Wit, J. An Input-Specific Orphan Receptor GPR158-HSPG Interaction Organizes Hippocampal Mossy Fiber-CA3 Synapses. Neuron 2018, 100, 201–215. [Google Scholar] [CrossRef] [Green Version]
- Orlandi, C.; Omori, Y.; Wang, Y.; Cao, Y.; Ueno, A.; Roux, M.J.; Condomitti, G.; de Wit, J.; Kanagawa, M.; Furukawa, T.; et al. Transsynaptic Binding of Orphan Receptor GPR179 to Dystroglycan-Pikachurin Complex. Is Essential for the Synaptic Organization of Photoreceptors. Cell Rep. 2018, 25, 130–145. [Google Scholar] [CrossRef] [Green Version]
- Khrimian, L.; Obri, A.; Ramos-Brossier, M.; Rousseaud, A.; Moriceau, S.; Nicot, A.S.; Mera, P.; Kosmidis, S.; Karnavas, T.; Saudou, F.; et al. Gpr158 mediates osteocalcin's regulation of cognition. J. Exp. Med. 2017, 214, 2859–2873. [Google Scholar] [CrossRef] [PubMed]
- Binet, V.; Brajon, C.; Le Corre, L.; Acher, F.; Pin, J.P.; Prézeau, L. The heptahelical domain of GABA(B2) is activated directly by CGP7930, a positive allosteric modulator of the GABA(B) receptor. J. Biol. Chem. 2004, 279, 29085–29091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binet, V.; Duthey, B.; Lecaillon, J.; Vol, C.; Quoyer, J.; Labesse, G.; Pin, J.P.; Prézeau, L. Common structural requirements for heptahelical domain function in class A and class C G protein-coupled receptors. J. Biol. Chem. 2007, 282, 12154–12163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galvez, T.; Duthey, B.; Kniazeff, J.; Blahos, J.; Rovelli, G.; Bettler, B.; Prézeau, L.; Pin, J.P. Allosteric interactions between GB1 and GB2 subunits are required for optimal GABA(B) receptor function. EMBO J. 2001, 20, 2152–2159. [Google Scholar] [CrossRef] [PubMed]
- Moreno Delgado, D.; Møller, T.C.; Ster, J.; Giraldo, J.; Maurel, D.; Rovira, X.; Scholler, P.; Zwier, J.M.; Perroy, J.; Durroux, T.; et al. Pharmacological evidence for a metabotropic glutamate receptor heterodimer in neuronal cells. eLife 2017, 6, e25233. [Google Scholar] [CrossRef] [PubMed]
- Levitz, J.; Habrian, C.; Bharill, S.; Fu, Z.; Vafabakhsh, R.; Isacoff, E.Y. Mechanism of Assembly and Cooperativity of Homomeric and Heteromeric Metabotropic Glutamate Receptors. Neuron 2016, 92, 143–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, Y.; Bush, M.; Mosyak, L.; Wang, F.; Fan, Q.R. Structural mechanism of ligand activation in human GABA(B) receptor. Nature 2013, 504, 254–259. [Google Scholar] [CrossRef] [Green Version]
- Nuemket, N.; Yasui, N.; Kusakabe, Y.; Nomura, Y.; Atsumi, N.; Akiyama, S.; Nango, E.; Kato, Y.; Kaneko, M.K.; Takagi, J.; et al. Structural basis for perception of diverse chemical substances by T1r taste receptors. Nat. Commun. 2017, 8, 15530. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Wang, C.; Gregory, K.J.; Han, G.W.; Cho, H.P.; Xia, Y.; Niswender, C.M.; Katritch, V.; Meiler, J.; Cherezov, V.; et al. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 2014, 344, 58–64. [Google Scholar] [CrossRef] [Green Version]
- Doré, A.S.; Okrasa, K.; Patel, J.C.; Serranovega, M.J.; Bennett, K.A.; Cooke, R.M.; Errey, J.C.; Jazayeri, A.; Khan, S.; Tehan, B.; et al. Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 2014, 511, 557–562. [Google Scholar] [CrossRef]
- Koehl, A.; Hu, H.; Feng, D.; Sun, B.; Zhang, Y.; Robertson, M.J.; Chu, M.; Kobilka, T.S.; Laeremans, T.; Steyaert, J.; et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 2019, 566, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Papasergi-Scott, M.M.; Robertson, M.J.; Seven, A.; Panova, O.; Mathiesen, J.M.; Skiniotis, G. Structures of metabotropic GABAB receptor. Nature 2020, 584, 310–314. [Google Scholar] [CrossRef] [PubMed]
- Shaye, H.; Ishchenko, A.; Lam, J.H.; Han, G.W.; Xue, L.; Rondard, P.; Pin, J.-P.; Katritch, V.; Gati, C.; Cherezov, V. Structural basis of the activation of a metabotropic GABA receptor. Nature 2020, 584, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.N.; Singh, S.; Laboute, T.; Strutzenberg, T.S.; Qiu, X.; Wu, D.; Novick, S.J.; Robinson, C.V.; Griffin, P.R.; Hunt, J.F.; et al. Cryo-EM structure of human GPR158 receptor coupled to the RGS7-Gβ5 signaling complex. Science 2021, 375, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Jeong, E.; Kim, Y.; Jeong, J.; Cho, Y. Structure of the class C orphan GPCR GPR158 in complex with RGS7-Gβ5. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Fini, M.E.; Schwartz, S.G.; Gao, X.; Jeong, S.; Patel, N.; Itakura, T.; Price, M.O.; Price, F.W.; Varma, R.; Stamer, W.D. Steroid-induced ocular hypertension/glaucoma: Focus on pharmacogenomics and implications for precision medicine. Prog. Retin. Eye Res. 2017, 56, 58–83. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; I Hurwitz, D.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; Farrens, D.L.; Altenbach, C.; Farahbakhsh, Z.T.; Hubbell, W.L.; Khorana, H.G. Structure and function in rhodopsin. Cysteines 65 and 316 are in proximity in a rhodopsin mutant as indicated by disulfide formation and interactions between attached spin labels. Biochemistry 1996, 35, 14040–14046. [Google Scholar] [CrossRef]
- Fay, J.F.; Dunham, T.D.; Farrens, D.L. Cysteine residues in the human cannabinoid receptor: Only C257 and C264 are required for a functional receptor, and steric bulk at C386 impairs antagonist SR141716A binding. Biochemistry 2005, 44, 8757–8769. [Google Scholar] [CrossRef]
- Agrawal, P.; Yu, K.; Salomon, A.R.; Sedivy, J.M. Proteomic profiling of Myc-associated proteins. Cell Cycle 2010, 9, 4908–4921. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto-Sato, E.; Fujimori, S.; Ishizaka, M.; Hirai, N.; Masuoka, K.; Saito, R.; Ozawa, Y.; Hino, K.; Washio, T.; Tomita, M.; et al. A comprehensive resource of interacting protein regions for refining human transcription factor networks. PLoS ONE 2010, 5, e9289. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Pyun, W.Y.; Park, H.W. Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets. Cells 2020, 9, 2308. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Sun, Z.; Sweat, Y.; Sweat, M.; Venugopalan, S.R.; Eliason, S.; Cao, H.; Paine, M.L.; Amendt, B.A. Pitx2-Sox2-Lef1 interactions specify progenitor oral/dental epithelial cell signaling centers. Development 2020, 147, dev186023. [Google Scholar] [CrossRef] [PubMed]
- Watkins, L.R.; Orlandi, C. In vitro profiling of orphan G protein coupled receptor (GPCR) constitutive activity. Br. J. Pharmacol. 2021, 178, 2963–2975. [Google Scholar] [CrossRef]
- Orlandi, C.; Xie, K.; Masuho, I.; Fajardo-Serrano, A.; Lujan, R.; Martemyanov, K.A. Orphan Receptor GPR158 Is an Allosteric Modulator of RGS7 Catalytic Activity with an Essential Role in Dictating Its Expression and Localization in the Brain. J. Biol. Chem. 2015, 290, 13622–13639. [Google Scholar] [CrossRef] [Green Version]
- Plattner, F.; Bibb, J.A. Chapter 25—Serine and Threonine Phosphorylation. In Basic Neurochemistry, 8th ed.; Brady, S.T., Siegel, G.J., Albers, R.W., Price, D.L., Eds.; Academic Press: New York, NY, USA, 2012; pp. 467–492. [Google Scholar] [CrossRef]
- Fernando, P.; Brunette, S.; Megeney, L.A. Neural stem cell differentiation is dependent upon endogenous caspase 3 activity. Faseb J. 2005, 19, 1671–1673. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Wang, L.; Xie, Z.; Zeng, C.; Shu, K. Transcriptomics Evidence for Common Pathways in Human Major Depressive Disorder and Glioblastoma. Int. J. Mol. Sci. 2018, 19, 234. [Google Scholar] [CrossRef] [Green Version]
- Koroknai, V.; Szász, I.; Hernandez-Vargas, H.; Fernandez-Jimenez, N.; Cuenin, C.; Herceg, Z.; Vízkeleti, L.; Ádány, R.; Ecsedi, S.; Balázs, M. DNA hypermethylation is associated with invasive phenotype of malignant melanoma. Exp. Dermatol. 2019, 29, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Oka, D.; Yamashita, S.; Tomioka, T.; Nakanishi, Y.; Kato, H.; Kaminishi, M.; Ushijima, T. The presence of aberrant DNA methylation in noncancerous esophageal mucosae in association with smoking history: A target for risk diagnosis and prevention of esophageal cancers. Cancer 2009, 115, 3412–3426. [Google Scholar] [CrossRef]
- Larouche, V.; Akirov, A.; AlShehri, S.; Ezzat, S. Management of Small Bowel Neuroendocrine Tumors. Cancers 2019, 11, 1395. [Google Scholar] [CrossRef] [Green Version]
- Bertani, E.; Ravizza, D.; Milione, M.; Massironi, S.; Grana, C.M.; Zerini, D.; Piccioli, A.N.; Spinoglio, G.; Fazio, N. Neuroendocrine neoplasms of rectum: A management update. Cancer Treat. Rev. 2018, 66, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Engqvist, H.; Parris, T.Z.; Kovács, A.; Rönnerman, E.W.; Sundfeldt, K.; Karlsson, P.; Helou, K. Validation of Novel Prognostic Biomarkers for Early-Stage Clear-Cell, Endometrioid and Mucinous Ovarian Carcinomas Using Immunohistochemistry. Front. Oncol. 2020, 10, 162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engqvist, H.; Parris, T.Z.; Kovács, A.; Nemes, S.; Rönnerman, E.W.; De Lara, S.; Biermann, J.; Sundfeldt, K.; Karlsson, P.; Helou, K. Immunohistochemical validation of COL3A1, GPR158 and PITHD1 as prognostic biomarkers in early-stage ovarian carcinomas. BMC Cancer 2019, 19, 928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chi, Q.; Zhao, Z. Up-regulation of long non-coding RNA SPRY4-IT1 promotes tumor cell migration and invasion in lung adenocarcinoma. Oncotarget 2017, 8, 51058–51065. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Wagner, F.; Bernard, R.; Derst, C.; French, L.; Veh, R.W. Microarray analysis of transcripts with elevated expressions in the rat medial or lateral habenula suggest fast GABAergic excitation in the medial habenula and habenular involvement in the regulation of feeding and energy balance. Anat. Embryol. 2016, 221, 4663–4689. [Google Scholar] [CrossRef]
- Piaggi, P.; Masindova, I.; Muller, Y.L.; Mercader, J.; Wiessner, G.B.; Chen, P.; SIGMA Type 2 Diabetes; Kobes, S.; Hsueh, W.-C.; Mongalo, M.; et al. A Genome-Wide Association Study Using a Custom Genotyping Array Identifies Variants in GPR158 Associated With Reduced Energy Expenditure in American Indians. Diabetes 2017, 66, 2284–2295. [Google Scholar] [CrossRef] [Green Version]
- Aïssani, B.; Perusse, L.; Lapointe, G.; Chagnon, Y.C.; Bouchard, L.; Walts, B.; Bouchard, C. A Quantitative Trait Locus for Body Fat on Chromosome 1q43 in French Canadians: Linkage and Association Studies. Obesity 2006, 14, 1605–1615. [Google Scholar] [CrossRef] [Green Version]
- Ferron, M.; Wei, J.; Yoshizawa, T.; Del Fattore, A.; DePinho, R.A.; Teti, A.; Ducy, P.; Karsenty, G. Insulin Signaling in Osteoblasts Integrates Bone Remodeling and Energy Metabolism. Cell 2010, 142, 296–308. [Google Scholar] [CrossRef] [Green Version]
- Ovsyannikova, I.G.; Kennedy, R.B.; O’Byrne, M.; Jacobson, R.M.; Pankratz, V.S.; Poland, G.A. Genome-wide association study of antibody response to smallpox vaccine. Vaccine 2012, 30, 4182–4189. [Google Scholar] [CrossRef] [Green Version]
- Vergara, C.; Thio, C.L.; Johnson, E.; Kral, A.H.; O’Brien, T.R.; Goedert, J.J.; Mangia, A.; Piazzolla, V.; Mehta, S.H.; Kirk, G.D.; et al. Multi-Ancestry Genome-Wide Association Study of Spontaneous Clearance of Hepatitis C Virus. Gastroenterology 2019, 156, 1496–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodder, E.M.; Scicluna, B.P.; Beekman, L.; Arends, D.; Moerland, P.D.; Tanck, M.W.; Adriaens, M.E.; Bezzina, C.R. Integrative Genomic Approach Identifies Multiple Genes Involved in Cardiac Collagen Deposition. Circ. Cardiovasc. Genet. 2014, 7, 790–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosmidis, S.; Polyzos, A.; Harvey, L.; Youssef, M.; Denny, C.A.; Dranovsky, A.; Kandel, E.R. RbAp48 Protein Is a Critical Component of GPR158/OCN Signaling and Ameliorates Age-Related Memory Loss. Cell Rep. 2018, 25, 959–973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mallah, K.; Quanico, J.; Raffo-Romero, A.; Cardon, T.; Aboulouard, S.; Devos, D.; Kobeissy, F.; Zibara, K.; Salzet, M.; Fournier, I. Mapping Spatiotemporal Microproteomics Landscape in Experimental Model of Traumatic Brain Injury Unveils a link to Parkinson’s Disease. Mol. Cell. Proteom. 2019, 18, 1669–1682. [Google Scholar] [CrossRef]
- Itakura, T.; Webster, A.; Chintala, S.K.; Wang, Y.; Gonzalez, J.M., Jr.; Tan, J.; Vranka, J.A.; Acott, T.; Craft, C.M.; Saber, M.E.S.; et al. GPR158 in the Visual System: Homeostatic Role in Regulation of Intraocular Pressure. J. Ocul. Pharmacol. Ther. 2019, 35, 203–215. [Google Scholar] [CrossRef]
- Fenner, A. Orphan receptor GPR158 finds a home in prostate cancer growth and progression. Nat. Rev. Urol. 2015, 12, 182. [Google Scholar] [CrossRef]
- Çetereisi, D.; Kramvis, I.; Gebuis, T.; van der Loo, R.J.; Gouwenberg, Y.; Mansvelder, H.D.; Li, K.W.; Smit, A.B.; Spijker, S. Gpr158 Deficiency Impacts Hippocampal CA1 Neuronal Excitability, Dendritic Architecture, and Affects Spatial Learning. Front. Cell. Neurosci. 2019, 13, 465. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Wang, Z.-Z.; Zhang, S.; Chu, S.-F.; Mou, Z.; Chen, N.-H. The effects of glucocorticoids on depressive and anxiety-like behaviors, mineralocorticoid receptor-dependent cell proliferation regulates anxiety-like behaviors. Behav. Brain Res. 2019, 362, 288–298. [Google Scholar] [CrossRef]
- Dumontet, T.; Hammer, G.D. Bones and adrenal organogenesis: How embryonic osteocalcin influences lifelong adrenal function. J. Clin. Investig. 2022, 132, e157200. [Google Scholar] [CrossRef]
- Yadav, V.K.; Berger, J.M.; Singh, P.; Nagarajan, P.; Karsenty, G. Embryonic osteocalcin signaling determines lifelong adrenal steroidogenesis and homeostasis in the mouse. J. Clin. Investig. 2022, 132, e153752. [Google Scholar] [CrossRef]
- Altmann, A.; Cash, D.M.; Bocchetta, M.; Heller, C.; Reynolds, R.; Moore, K.; Convery, R.S.; Thomas, D.L.; van Swieten, J.C.; Moreno, F.; et al. Analysis of brain atrophy and local gene expression in genetic frontotemporal dementia. Brain Commun. 2020, 2, fcaa122. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Jia, L.; Li, F.; Jia, J. Identification of KIAA0513 and Other Hub Genes Associated With Alzheimer Disease Using Weighted Gene Coexpression Network Analysis. Front. Genet. 2020, 11, 981. [Google Scholar] [CrossRef] [PubMed]
- Alsaleh, H.; Haddrill, P.R. Identifying blood-specific age-related DNA methylation markers on the Illumina MethylationEPIC® BeadChip. Forensic Sci. Int. 2019, 303, 109944. [Google Scholar] [CrossRef] [PubMed]
- Damjanoska, K.J.; Heidenreich, B.A.; Kindel, G.H.; d’Souza, D.N.; Zhang, Y.; Garcia, F.; Battaglia, G.; Wolf, W.A.; can de Kar, L.D.; Muma, N.A. Agonist-Induced Serotonin 2A Receptor Desensitization in the Rat Frontal Cortex and Hypothalamus. J. Pharmacol. Exp. Ther. 2004, 309, 1043–1050. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, T.; Tari, P.K.; Connor, S.A.; Zhang, P.; Dobie, F.; She, K.; Kawabe, H.; Wang, Y.T.; Brose, N.; Craig, A.M. An LRRTM4-HSPG Complex Mediates Excitatory Synapse Development on Dentate Gyrus Granule Cells. Neuron 2013, 79, 680–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oury, F.; Khrimian, L.; Denny, C.A.; Gardin, A.; Chamouni, A.; Goeden, N.; Huang, Y.-Y.; Lee, H.; Srinivas, P.; Gao, X.-B.; et al. Maternal and Offspring Pools of Osteocalcin Influence Brain Development and Functions. Cell 2013, 155, 228–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karsenty, G.; Olson, E.N. Bone and Muscle Endocrine Functions: Unexpected Paradigms of Inter-organ Communication. Cell 2016, 164, 1248–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, K.; Nabeshima, T. Brain-Derived Neurotrophic Factor/TrkB Signaling in Memory Processes. J. Pharmacol. Sci. 2003, 91, 267–270. [Google Scholar] [CrossRef] [Green Version]
- Rowe, W.B.; Blalock, E.M.; Chen, K.-C.; Kadish, I.; Wang, D.; Barrett, J.E.; Thibault, O.; Porter, N.M.; Rose, G.M.; Landfield, P.W. Hippocampal Expression Analyses Reveal Selective Association of Immediate-Early, Neuroenergetic, and Myelinogenic Pathways with Cognitive Impairment in Aged Rats. J. Neurosci. 2007, 27, 3098–3110. [Google Scholar] [CrossRef]
- Wu, J.; Dou, Y.; Liu, W.; Zhao, Y.; Liu, X. Osteocalcin improves outcome after acute ischemic stroke. Aging 2020, 12, 387–396. [Google Scholar] [CrossRef]
- Foster, S.R.; Hauser, A.S.; Vedel, L.; Strachan, R.T.; Huang, X.-P.; Gavin, A.C.; Shah, S.D.; Nayak, A.P.; Haugaard-Kedström, L.; Penn, R.B.; et al. Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors. Cell 2019, 179, 895–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, G.R.; Posokhova, E.; Martemyanov, K.A. The R7 RGS Protein Family: Multi-Subunit Regulators of Neuronal G Protein Signaling. Cell Biophys. 2009, 54, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sunahara, R.K.; Dessauer, C.W.; Gilman, A.G. Complexity and diversity of mammalian adenylyl cyclases. Annu. Rev. Pharmacol. Toxicol. 1996, 36, 461–480. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vives, M.V.; Barbero-Castillo, A.; Perez-Zabalza, M.; Reig, R. GABAB receptors: Modulation of thalamocortical dynamics and synaptic plasticity. Neuroscience 2020, 456, 131–142. [Google Scholar] [CrossRef]
- Ostrovskaya, O.I.; Orlandi, C.; Fajardo-Serrano, A.; Young, S.; Lujan, R.; Martemyanov, K.A. Inhibitory Signaling to Ion Channels in Hippocampal Neurons Is Differentially Regulated by Alternative Macromolecular Complexes of RGS7. J. Neurosci. 2018, 38, 10002–10015. [Google Scholar] [CrossRef]
- Song, C.; Orlandi, C.; Sutton, L.P.; Martemyanov, K.A. The signaling proteins GPR158 and RGS7 modulate excitability of L2/3 pyramidal neurons and control A-type potassium channel in the prelimbic cortex. J. Biol. Chem. 2019, 294, 13145–13157. [Google Scholar] [CrossRef]
- Watkins, L.R.; Orlandi, C. Orphan G Protein Coupled Receptors in Affective Disorders. Genes 2020, 11, 694. [Google Scholar] [CrossRef]
- Hörnberg, H.; Pérez-Garci, E.; Schreiner, D.; Hatstatt-Burklé, L.; Magara, F.; Baudouin, S.; Matter, A.; Nacro, K.; Pecho-Vrieseling, E.; Scheiffele, P. Rescue of oxytocin response and social behaviour in a mouse model of autism. Nature 2020, 584, 252–256. [Google Scholar] [CrossRef]
- Xiang, D.; Wang, H.; Sun, S.; Yao, L.; Li, R.; Zong, X.; Wang, G.; Liu, Z. GRP Receptor Regulates Depression Behavior via Interaction With 5-HT2a Receptor. Front. Psychiatry 2020, 10, 1020. [Google Scholar] [CrossRef] [Green Version]





| Disorders/Phenotypes | Results of Studies |
|---|---|
| Human studies: | |
| Major Depressive Disorder | ↑ GPR158 in dlPFC [18,19] |
| Animal studies: | |
| Stress-induced Depression | ↑ GPR158 in mPFC, under chronic PRS [18] |
| ↑ GPR158 in mPFC, under UCMS [18] | |
| ↑ GPR158 in mPFC, with chronic corticosterone treatment [18] | |
| ↑ GPR158 in primary cortical neurons, with chronic corticosterone treatment [18] | |
| GPR158 OE in mPFC↑ Immobility in FST [18] | |
| GPR158 KO ↓ Immobility in FST [18] | |
| GPR158 KO↓ Marble buried in MBT [18] | |
| GPR158 KO↑ Time in open arms in EMP, 2–4 month olds [18] | |
| GPR158 KO ↓ time in open arms in EMP, 3 month-old females [18] | |
| GPR158 KO ↓ time in lit box in LDT, 3 month-old females [31] | |
| GPR158 KO↓ time in center in OFT, 3 month-old females [31] | |
| GPR158 KO ↔ time in center in OFT, 8–12 week-old males [78] | |
| GPR158 KO ↔ immobility time after yohimbine injection in FST, 2–4 month-old males [19] | |
| GPR158 KO↓ immobility in TST, 2–4 month olds [18] | |
| GPR158 OE in mPFC↑ immobility in TST, 2–4 month olds [18] | |
| GPR158 KO ↔ immobility time in TST, after yohimbine injection (not after vehicle injection),2–4 month-old males [19] | |
| Age-related Memory Loss | Disrupted GPR158/OCN signaling in the hippocampus [74] |
| Impaired Spatial Learning | Disrupted CA1 morphology and impaired spatial memory acquisition, GPR158 global KO [55,78] |
| Presynaptic Differentiation | ↑ Mossy fiber synapse density, impaired postsynaptic density and synaptic strength, GPR158 global KO [29] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, X.; Wei, S.; Wang, T.; Fan, H.; Zhang, Y.; Costa, C.D.; Brandner, S.; Yang, G.; Pan, Y.; He, Y.; et al. Research Status of the Orphan G Protein Coupled Receptor 158 and Future Perspectives. Cells 2022, 11, 1334. https://doi.org/10.3390/cells11081334
Fu X, Wei S, Wang T, Fan H, Zhang Y, Costa CD, Brandner S, Yang G, Pan Y, He Y, et al. Research Status of the Orphan G Protein Coupled Receptor 158 and Future Perspectives. Cells. 2022; 11(8):1334. https://doi.org/10.3390/cells11081334
Chicago/Turabian StyleFu, Xianan, Shoupeng Wei, Tao Wang, Hengxin Fan, Ying Zhang, Clive Da Costa, Sebastian Brandner, Guang Yang, Yihang Pan, Yulong He, and et al. 2022. "Research Status of the Orphan G Protein Coupled Receptor 158 and Future Perspectives" Cells 11, no. 8: 1334. https://doi.org/10.3390/cells11081334