The Role of N-Glycosylation in the Intracellular Trafficking and Functionality of Neuronal Growth Regulator 1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. Plasmids and Site-Directed Mutagenesis
2.3. Immunoblotting and Antibodies
2.4. Homophilic Binding and Immunofluorescence Microscopy
2.5. Protein Deglycosylation
2.6. Cell Surface Biotinylation
2.7. Lipid Raft Fractionation and Cell Aggregation Assay
3. Results
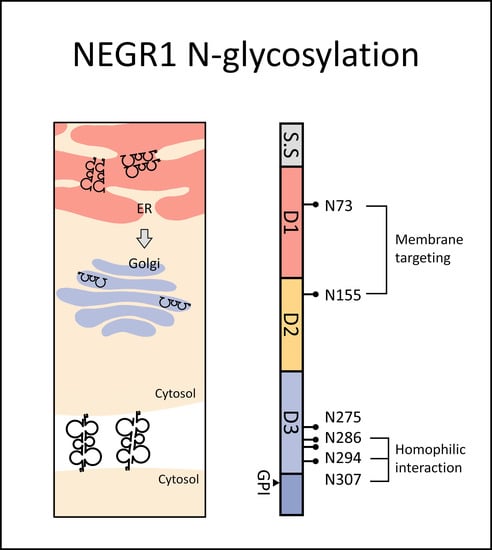
3.1. Glycosylation Status of NEGR1 Protein
3.2. Analysis of NEGR1 N-Glycosylation Mutants
3.3. Membrane Localization of the NEGR1 Mutant Proteins
3.4. Subcellular Localization of NEGR1 Mutants
3.5. Characterization of NEGR1 N-Glycosylation Mutants
3.6. Homophilic Interaction of NEGR1 N-Glycosylation Mutants
3.7. Cell Aggregation Assay and Lipid Raft Fractionation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Abu-Qarn, M.; Eichler, J.; Sharon, N. Not just for Eukarya anymore: Protein glycosylation in Bacteria and Archaea. Curr. Opin. Struct. Biol. 2008, 18, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Ioffe, E.; Stanley, P. Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc. Natl. Acad. Sci. USA 1994, 91, 728–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scott, D.W.; Tolbert, C.E.; Graham, D.M.; Wittchen, E.; Bear, J.E.; Burridge, K. N-glycosylation controls the function of junctional adhesion molecule-A. Mol. Biol. Cell 2015, 26, 3205–3214. [Google Scholar] [CrossRef] [PubMed]
- Freeze, H.H.; Eklund, E.A.; Ng, B.G.; Patterson, M.C. Neurological aspects of human glycosylation disorders. Annu. Rev. Neurosci. 2015, 38, 105–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleene, R.; Schachner, M. Glycans and neural cell interactions. Nat. Rev. Neurosci. 2004, 5, 195–208. [Google Scholar] [CrossRef]
- Freeze, H.H.; Eklund, E.A.; Ng, B.G.; Patterson, M.C. Neurology of inherited glycosylation disorders. Lancet Neurol. 2012, 11, 453–466. [Google Scholar] [CrossRef] [Green Version]
- Hildebrandt, H.; Muhlenhoff, M.; Weinhold, B.; Gerardy-Schahn, R. Dissecting polysialic acid and NCAM functions in brain development. J. Neurochem. 2007, 103 (Suppl. 1), 56–64. [Google Scholar] [CrossRef]
- Funatsu, N.; Miyata, S.; Kumanogoh, H.; Shigeta, M.; Hamada, K.; Endo, Y.; Sokawa, Y.; Maekawa, S. Characterization of a novel rat brain glycosylphosphatidylinositol-anchored protein (Kilon), a member of the IgLON cell adhesion molecule family. J. Biol. Chem. 1999, 274, 8224–8230. [Google Scholar] [CrossRef] [Green Version]
- Szczurkowska, J.; Pischedda, F.; Pinto, B.; Manago, F.; Haas, C.A.; Summa, M.; Bertorelli, R.; Papaleo, F.; Schafer, M.K.; Piccoli, G.; et al. NEGR1 and FGFR2 cooperatively regulate cortical development and core behaviours related to autism disorders in mice. Brain J. Neurol. 2018, 141, 2772–2794. [Google Scholar] [CrossRef]
- Veerappa, A.M.; Saldanha, M.; Padakannaya, P.; Ramachandra, N.B. Family-based genome-wide copy number scan identifies five new genes of dyslexia involved in dendritic spinal plasticity. J. Hum. Genet. 2013, 58, 539–547. [Google Scholar] [CrossRef] [Green Version]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014, 511, 421–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, H.; Xu, M.; Zhan, G.L.; Fan, Y.; Zhou, H.; Jiang, H.Y.; Lu, W.H.; Tan, L.; Zhang, D.F.; Yao, Y.G.; et al. The GWAS Risk Genes for Depression May Be Actively Involved in Alzheimer’s Disease. J. Alzheimer’s Dis. JAD 2018, 64, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Hyde, C.L.; Nagle, M.W.; Tian, C.; Chen, X.; Paciga, S.A.; Wendland, J.R.; Tung, J.Y.; Hinds, D.A.; Perlis, R.H.; Winslow, A.R. Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat. Genet. 2016, 48, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Noh, K.; Lee, H.; Choi, T.Y.; Joo, Y.; Kim, S.J.; Kim, H.; Kim, J.Y.; Jahng, J.W.; Lee, S.; Choi, S.Y.; et al. Negr1 controls adult hippocampal neurogenesis and affective behaviors. Mol. Psychiatry 2019, 24, 1189–1205. [Google Scholar] [CrossRef]
- Joo, Y.; Kim, H.; Lee, S.; Lee, S. Neuronal growth regulator 1-deficient mice show increased adiposity and decreased muscle mass. Int. J. Obes. 2019, 43, 1769–1782. [Google Scholar] [CrossRef]
- Kim, H.; Chun, Y.; Che, L.; Kim, J.; Lee, S.; Lee, S. The new obesity-associated protein, neuronal growth regulator 1 (NEGR1), is implicated in Niemann-Pick disease Type C (NPC2)-mediated cholesterol trafficking. Biochem. Biophys. Res. Commun. 2017, 482, 1367–1374. [Google Scholar] [CrossRef]
- Kim, H.; Hwang, J.S.; Lee, B.; Hong, J.; Lee, S. Newly Identified Cancer-Associated Role of Human Neuronal Growth Regulator 1 (NEGR1). J. Cancer 2014, 5, 598–608. [Google Scholar] [CrossRef] [Green Version]
- Cheon, Y.; Yoo, A.; Seo, H.; Yun, S.Y.; Lee, H.; Lim, H.; Kim, Y.; Che, L.; Lee, S. Na/K-ATPase beta1-subunit associates with neuronal growth regulator 1 (NEGR1) to participate in intercellular interactions. BMB Rep. 2021, 54, 164–169. [Google Scholar] [CrossRef]
- Marinko, J.T.; Huang, H.; Penn, W.D.; Capra, J.A.; Schlebach, J.P.; Sanders, C.R. Folding and Misfolding of Human Membrane Proteins in Health and Disease: From Single Molecules to Cellular Proteostasis. Chem. Rev. 2019, 119, 5537–5606. [Google Scholar] [CrossRef]
- Ranaivoson, F.M.; Turk, L.S.; Ozgul, S.; Kakehi, S.; von Daake, S.; Lopez, N.; Trobiani, L.; De Jaco, A.; Denissova, N.; Demeler, B.; et al. A Proteomic Screen of Neuronal Cell-Surface Molecules Reveals IgLONs as Structurally Conserved Interaction Modules at the Synapse. Structure 2019, 27, 893–906.e9. [Google Scholar] [CrossRef]
- Wang, B.; Stanford, K.R.; Kundu, M. ER-to-Golgi Trafficking and Its Implication in Neurological Diseases. Cells 2020, 9, 408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanus, C.; Geptin, H.; Tushev, G.; Garg, S.; Alvarez-Castelao, B.; Sambandan, S.; Kochen, L.; Hafner, A.S.; Langer, J.D.; Schuman, E.M. Unconventional secretory processing diversifies neuronal ion channel properties. Elife 2016, 5, e20609. [Google Scholar] [CrossRef] [PubMed]
- Bowen, A.B.; Bourke, A.M.; Hiester, B.G.; Hanus, C.; Kennedy, M.J. Golgi-independent secretory trafficking through recycling endosomes in neuronal dendrites and spines. Elife 2017, 6, e27362. [Google Scholar] [CrossRef] [PubMed]
- Govind, A.P.; Jeyifous, O.; Russell, T.A.; Yi, Z.; Weigel, A.V.; Ramaprasad, A.; Newell, L.; Ramos, W.; Valbuena, F.M.; Casler, J.C.; et al. Activity-dependent Golgi satellite formation in dendrites reshapes the neuronal surface glycoproteome. Elife 2021, 10, e68910. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Jayaram, M.; Kaare, M.; Leidmaa, E.; Jagomae, T.; Heinla, I.; Hickey, M.A.; Kaasik, A.; Schafer, M.K.; Innos, J.; et al. Neural cell adhesion molecule Negr1 deficiency in mouse results in structural brain endophenotypes and behavioral deviations related to psychiatric disorders. Sci. Rep. 2019, 9, 5457. [Google Scholar] [CrossRef]
- Itoh, S.; Hachisuka, A.; Kawasaki, N.; Hashii, N.; Teshima, R.; Hayakawa, T.; Kawanishi, T.; Yamaguchi, T. Glycosylation analysis of IgLON family proteins in rat brain by liquid chromatography and multiple-stage mass spectrometry. Biochemistry 2008, 47, 10132–10154. [Google Scholar] [CrossRef]
- Venkannagari, H.; Kasper, J.M.; Misra, A.; Rush, S.A.; Fan, S.; Lee, H.; Sun, H.; Seshadrinathan, S.; Machius, M.; Hommel, J.D.; et al. Highly Conserved Molecular Features in IgLONs Contrast Their Distinct Structural and Biological Outcomes. J. Mol. Biol. 2020, 432, 5287–5303. [Google Scholar] [CrossRef]
- Phillips, B.P.; Gomez-Navarro, N.; Miller, E.A. Protein quality control in the endoplasmic reticulum. Curr. Opin. Cell. Biol. 2020, 65, 96–102. [Google Scholar] [CrossRef]
- Scott, H.; Panin, V.M. N-glycosylation in regulation of the nervous system. Adv. Neurobiol. 2014, 9, 367–394. [Google Scholar] [CrossRef] [Green Version]







| Mutant Name | Mutation Sites |
|---|---|
| Single mutant | N73Q, N155Q, N275Q, N286Q, N294Q, or N307Q |
| N73Q/N155Q | N73Q + N155Q |
| 2MT | N286Q + N294Q |
| 3MT | N286Q + N294Q + N307Q |
| 4MT | N286Q + N294Q + N307Q + N275Q |
| 5MT | N286Q + N294Q + N307Q + N275Q + N155Q |
| 6MT | N286Q + N294Q + N307Q + N275Q + N155Q + N73Q |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sim, G.; Jeong, M.; Seo, H.; Kim, J.; Lee, S. The Role of N-Glycosylation in the Intracellular Trafficking and Functionality of Neuronal Growth Regulator 1. Cells 2022, 11, 1242. https://doi.org/10.3390/cells11071242
Sim G, Jeong M, Seo H, Kim J, Lee S. The Role of N-Glycosylation in the Intracellular Trafficking and Functionality of Neuronal Growth Regulator 1. Cells. 2022; 11(7):1242. https://doi.org/10.3390/cells11071242
Chicago/Turabian StyleSim, Gyuri, Moonkyung Jeong, Hyunseok Seo, Jangrae Kim, and Soojin Lee. 2022. "The Role of N-Glycosylation in the Intracellular Trafficking and Functionality of Neuronal Growth Regulator 1" Cells 11, no. 7: 1242. https://doi.org/10.3390/cells11071242