2. Materials and Methods
2.1. PCR-Based Cloning
The component fragments were PCR-amplified using Q5 High-Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA, USA) with a 65 °C annealing temperature.
2.2. Colony PCR
Colony PCR was performed using GoTaq Master Mixes (Promega, Madison, WI, USA) with a 55 °C annealing temperature.
2.3. Restriction Digest of Plasmid DNA
The destination plasmid DNA was digested using BsaI-HFv2 (New England Biolabs, Ipswich, MA, USA). This step is useful to increase ligation efficiency for gRNA array with more than two gRNAs.
2.4. Gel Purification
The PCR products and digested destination vectors were purified using the Zymoclean Gel DNA Recovery Kit (ZYMO RESEARCH, Irvine, CA, USA).
2.5. Golden Gate Assembly
Assembly reactions were performed in a thermocycler using the NEBridge Golden Gate Assembly Kit (BsaI-HFv2) (New England Biolabs, Ipswich, MA, USA) with the suggested assembly protocol.
2.6. Plasmid Sequencing
The plasmids were Sanger-sequenced using SimpleSeq Kit Premixed (Eurofins Genomics, Louisville, KY, USA). The sequencing data were aligned with a plasmid sequence in SnapGene.
2.7. E. coli Transformation
The E. coli transformation was performed using NEB 5-alpha Competent E. coli (New England Biolabs, Ipswich, MA, USA), following the manual.
2.8. Plasmid Isolation
The plasmid DNA extraction was performed using GenElute Plasmid Miniprep Kit (Sigma-Aldrich, Saint Louis, MO, USA).
2.9. Oligos Annealing
The two oligo strands were added together in equal molar amounts. The mixed oligonucleotides were heated to 94 °C for 2 min, then gradually cooled.
2.10. Vector Cloning
The U6 promoter in the pKSE401 vector was replaced by a U3 promoter via NEBuilder HiFi DNA Assembly (New England Biolabs, Ipswich, MA, USA), and a window sequence (GGTCGGAGACCAACGGTCTCGGTGGCACCGAGTCGGTGCTTTTTTT) was inserted between the U3 promoter and its terminator. The template vectors were generated by inserting two gBlocks Gene Fragments (IDT, Coralville, IA, USA) into a modified pKSE401 vector via NEBuilder HiFi DNA Assembly. Information for all primers and gBlocks used in this study is provided in
Table S1.
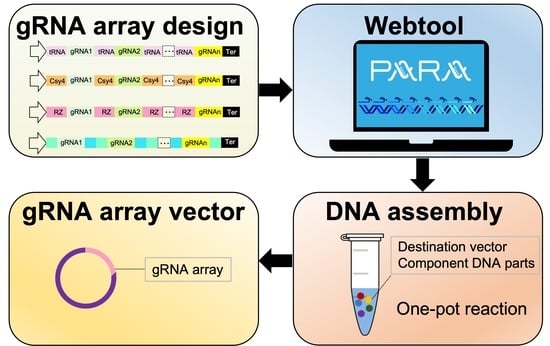
2.11. Web Tool Design
PARAweb is a web tool that provides a complete workflow for the design and assembly of gRNA arrays for multiplex genome editing. PARAweb features a series of drop-down menus that the user may interact with to choose the parameters for the design tool. Parameters include the type of multi-gRNA expression system, the ligation action, the appropriate restriction enzyme and the organism type. After selecting parameters, the user uploads a file containing the gRNA sequences of the gRNA array. The OHs are chosen via algorithm (
Figure 1), and a list of primers is displayed in tabular, color-coded format for the PCR amplification of DNA fragments. When the complete sequences are downloaded, DNA constants relevant to specific gRNA mode sets are used. The resulting text files contain the primers, the component DNA fragments of the gRNA array and the complete gRNA array assembly sequence.
Figure 2 illustrates the workflow implemented in PARAweb.
For steps 1 and 2, we created the interface of PARAweb, including the name, featured figure, drop-down menus and upload zone. When the defined gRNA sequences and destination vector sequences are given in step 1 and 2, to select high-fidelity OH sets, step 3 is performed for global optimization of OHs from gRNA sequences via: (a) identification of candidate OHs from each of the 20-nt gRNA sequences; (b) identification of all OH combinations with a pairwise crossmatch score <30 from identified candidate OHs in step (a); and (c) identification of the best OH combination with the highest total self-match score for assembling the gRNA array, as illustrated in
Figure 1. The crossmatch score and self-match score were used based on the comprehensive profiling of 4 base OH ligation fidelity by T4 DNA ligase [
6,
7].
Once the OH is selected for each gRNA sequence, the required oligos/primers are generated in step 4. For each primer, the 5′ end of a template-specific sequence is flanked in an orderly manner by 1 BsaI restriction site, 1 specific 4-bp OH sequence and 1 gRNA sequence. In step 5, each component DNA fragment is generated by combining the corresponding forward primer (F [n]), predefined template sequence, and reverse primer (R [n]). In step 6, the assembled gRNA array sequence is generated by combining individual component DNA fragments from step 5. In step 7, assembled vector sequences are generated by connecting the user-provided destination vector and assembled gRNA array sequence from step 6. In step 8, all the described outputs, including the required oligos (step 4), component DNA fragments (step 5), assembled gRNA array sequence (step 6) and assembled vector sequences (step 7), can be downloaded as individual text files.
3. Results
Generally, directly synthesizing gRNA arrays is challenging because of their highly repetitive elements. Inspired by the multiplexed genome editing with the endogenous transfer RNA (tRNA)–processing system in rice [
8], we developed the PCR-based PARA method for the assembly of tRNA-gRNA arrays using Golden Gate (GG) assembly (
Figure 3a). To assemble, in an orderly manner, multiple fragments simultaneously, the fragment-specific sequences of 4-base overhangs (OHs) are an essential prerequisite. Unlike the modular cloning with predefined OHs, in the PARA method, the 4-bp OHs are selected from distinct gRNA sequences. Therefore, no scar sequences are introduced during cloning. Thus, the gRNA arrays can be divided into multiple individual DNA parts. Each of the DNA parts can be generated through PCR amplification of a predesigned template vector (
Figure 3a). Furthermore, [n + 1] fragments can be used for the assembly of [n] gRNAs. Next, the DNA fragments are ligated, in an orderly manner, into a destination vector containing two predesigned BsaI restriction sites to form a gRNA array within an expression vector (
Figure 3a).
One critical step in the PARA method is the design of required oligos (i.e., primers) for the PCR amplification of component fragments. For each primer, the 5′ end of a template-specific sequence is flanked, in an orderly manner, by 1 BsaI restriction site, 1 specific 4-bp OH sequence and 1 gRNA sequence (
Figure 3b). Two OHs in the first forward primer and last reverse primer must be complementary with the sticky end of the destination vector digested by BsaI. Moreover, all selected OHs must be distinctive, with low similarity to one another to ensure the orderly assembly of gRNA arrays.
Using the PARA method, the expression vector containing a gRNA array can be constructed within three days, which is, to date, the fastest method for the assembly of gRNA arrays (
Figure 3c), saving up to 70% of time and effort in comparison with traditional methods [
9,
10,
11,
12]. Depending on the user preference and project requirements, the component DNA fragments can also be generated using commercial DNA synthesis or via annealing long oligos, allowing for high-throughput library synthesis.
To explore the capacity of the PARA method, we performed multi-gRNA assembly with various numbers of gRNAs using the plant tRNA-gRNA system. Four target genes of
Populus deltoides WV94 were selected from Phytozome [
13], and five gRNAs were designed for each gene using a gRNA design web tool, CHOPCHOP [
14]. Required oligonucleotides were designed manually as illustrated in
Figure 3b. The component fragments were generated through PCR amplification of the predesigned template vector type I followed by gel purification (
Figures S1a and S2;
Table S2). Then, all component fragments were assembled into a modified pKSE401 vector [
15], followed by transformation on day 1. Numerous colonies were observed on the selection medium on day 2 (
Figure S3a). Next, we analyzed the colonies via colony PCR (
Figure S4) and Sanger sequencing. As expected, the efficiency of GG assembly gradually decreased with the increase in the total number of gRNAs (
Figure 4a). In 2-gRNA assembly, target bands were observed in all selected colonies (
n = 18) (
Figure S4a). When the number of gRNAs exceeded 2, false positive colonies were detected on the selection medium (
Figure S4b–i). Interestingly, in 4-gRNA assembly, 90% of the transformants harbored correctly assembled constructs (
Figure 4a). In the assembly with between 6 and 10 gRNAs, the positive rate of transformants ranged from ~50% to ~80%. To explore the potential of the PARA method, we further studied the assembly of gRNA arrays with up to 20 gRNAs. More than 25% of the analyzed transformants contained the correctly assembled constructs when the number of gRNAs was under 16, and the positive rate decreased to below 10% when the number of gRNAs was 16 or more (
Figure 4a). Two transformants were randomly selected from each replicate and ordered. The orientation of the constructs was verified using Sanger sequencing. Overall, we demonstrate through this method that the PARA method is an effective approach for the one-pot assembly of gRNA arrays with up to 16 gRNAs.
In addition to the tRNA-gRNA system, polycistronic transcripts can also be processed post-transcriptionally into individual gRNAs by other RNA-cleaving enzymes, such as the CRISPR-associated RNA endoribonuclease Csy4 [
16] and ribozymes (RBs) [
17]. Recently, multiplexed CRISPR/Cas9 genome editing has been successfully applied in yeast [
18], human cells [
19] and plants [
5]. We tested the PARA method for the assembly of gRNA arrays based on Csy4 and RB expression systems, and we compared the cloning efficiency of gRNA arrays containing the same set of eight gRNAs in different gRNA expression systems based on tRNA, Csy4 and RB (
Figure 4b,c ). All component DNA fragments were generated by either PCR amplification of the predesigned template vector type I or annealing oligonucleotides. High-efficiency cloning was achieved in the Csy4 system (80.0%), tRNA system (73.4%) and RB system (63.0%) (
Figure 4c). Recently, it was reported that multiplexed CRISPR/Cas12a was able to target multiple sites with high biallelic editing efficiency in rice using the processing system of the hammerhead (HH) and hepatitis delta virus (HDV) RBs (
Figure 4b) [
9]. However, the assembly of such a sophisticated construct is difficult and time-consuming. In this study, we sought to create gRNA arrays containing the same components in a one-step effort using the PARA method. Required oligonucleotides were designed manually using the same strategy as shown in
Figure 3b. Component fragments were generated through PCR amplification using a predesigned template vector type II (
Figure S1b and
Table S2). In the 8-gRNA assembly, approximately 36.4% of the analyzed transformants contained the correctly assembled construct (
Figure 4c ). Other than PCR amplicons, the HH-HDV-RB array with eight gRNAs was also assembled successfully using synthesized DNA fragments (
Figures S3e and S5). Altogether, the PARA method is a potent and robust approach for assembling gRNA arrays with different expression systems.
Based on the literature, multiple tRNA systems with organism-specific tRNA sequences have been used in plants [
5,
8], yeast [
3] and
Drosophila [
4]. The Csy4 system has been used in plants [
5], yeast [
18] and human cells [
19]. The RB and HH-HDV-RB systems have also been used in plants [
5,
9]. To simplify vector design and construction, we developed a dedicated web tool, PARAweb, which allows users to accurately design and simulate complex cloning procedures that involve numerous gRNAs. PARAweb can be freely accessed at
https://fair.ornl.gov/BioDesign/para/para/ (accessed on 17 June 2022). The PARAweb tool is suitable for the design of all of the described gRNA array expression systems (i.e., tRNA, Csy4 and RB for Cas9, as well as HH-HDV-RB for Cas12a) (
Figure 4d). Moreover, this web-based gRNA array tool is useful for the application of multiplexed CRISPR knockout, base editing, CRISPRa and CRISPRi in a wide variety of organisms, including animals, plants and microbes. With given input gRNA sequences, PARAweb can generate PCR primers, component fragments and linear assembled gRNA array sequences (
Figure 4e). When a valid destination vector sequence is given, PARAweb can also generate the assembled vector sequences containing the gRNA array (
Figure 4e ). Notably, the ligation frequency for each OH pair in assembly reactions with BsaI-HFv2 and T4 DNA ligase [
7] is utilized as a basic rule to select high-fidelity OH sets in PARAweb. Eight poplar gRNAs used previously were selected to test PARAweb, generating PCR primers, component fragments, linear assembled gRNA array sequences and assembled vector sequences (
Figure S6 and
Table S3). These component fragments were successfully PCR-amplified with the primers and ligated through linear ligation or cloned into a modified pKSE401 vector in SnapGene (
Figure S7). Following the procedures described in
Figure 3, we detected 55.6% positive colonies in three biological replicates (
Figures S3f and S8).