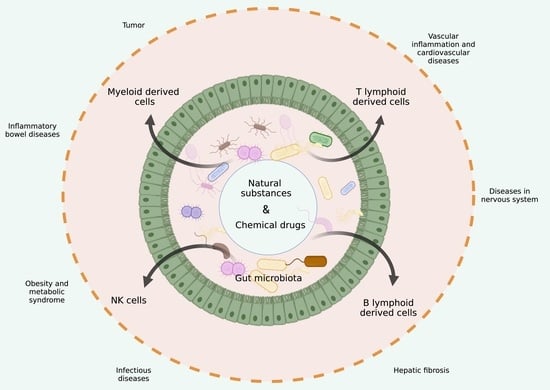
Gut Microbiota-Derived Tryptophan Metabolites Maintain Gut and Systemic Homeostasis
Abstract
:1. Introduction
2. Trp Metabolism in Gut Microbiota
2.1. Trp Metabolism in Gut Microbiota
2.2. Regulation of Trp Metabolism in Gut Microbiota
3. Regulation of Trp Metabolites from Gut Microbiota in the Immune Cells
3.1. Macrophages
- (1)
- Inflammatory and immunosuppressive macrophages. In vitro studies show that Trp metabolites suppress inflammatory responses in macrophages. Trp metabolite receptor AhR signaling has an important role in the function of macrophages [64]. AhR is required for the amelioration of Streptococcus and Salmonella typhimurium-induced immunopathology in LPS-tolerant mice [65]. AhR knockout (KO) mice were more sensitive to LPS-induced lethal shock [66]. These AhR KO mice can produce higher amounts of proinflammatory cytokines (TNF, IL-6 and IL-12). Others also found that the decreased inflammatory processes in LPS-activated macrophages by an endogenous (FICZ) or exogenous (BaP) ligand is partially dependent on AhR signaling [67]. AhR downregulates the production of the proinflammatory cytokine IL-6 through suppressing histamine production in macrophages [68]. The immunomodulatory roles of AhR are through a Rac1 ubiquitination-dependent mechanism, which can attenuate AKT signaling and result in a mitigated inflammatory response [67]. The proteomic changes in the macrophages after treatment with Trp metabolites I3AA or IAld, as well as AhR ligand benzo(a) pyrene (BaP), showed that fatty acid β-oxidation and oxidative phosphorylation were significantly increased in a time- and LPS-dependent manner [69]. In addition, the AhR-Src-STAT3-IL-10 signaling pathway is also a critical pathway in regulating inflammatory macrophages [70].
- (2)
- Gut resident macrophages. It is unclear how Trp metabolites regulate these resident macrophages. In gut tissues, Muller et al. [74] discussed the origin, phenotype and function of the resident macrophages in the different layers of the intestines during homeostasis. A “monocyte waterfall” from circulation to the intestine maintains the macrophage pool in a CCR2-dependent manner in the murine colon [75]. The monocytes, which are identified as the ly6chi CX3CR1int MHCII− subset, exhibit proinflammatory properties. They terminally differentiate into mature resident ly6clow/−CX3CR1hiMHCIIhi macrophages in the intestines. The resident macrophages reside in the lamina propria (LP) or the muscle layer. LP macrophages (LPMs) may be subdivided into mucosal and submucosal LPMs [76], whereas mucosal LPMs line the intestinal epithelium and vasculature in the intestines [77,78]. These macrophages contribute to the host defense, barrier integrity and constitutive secretion of interleukin (IL)-10, which can promote the maintenance of FoxP3+ Treg [79]. Perivascular macrophages participate in the regulation of the vasculature in the small intestine and colon [76,78]. The macrophages residing in the muscularis are essential for tissue homeostasis. Their interactions in muscularis macrophages with the neurons control intestinal motility and protect tissues. These macrophage subsets in gut tissues belong to resident macrophages (anti-inflammatory macrophages). However, how Trp metabolites regulate these resident macrophages remains unclear.
- (3)
- Myeloid precursor cells. The activation of Trp metabolites receptor AhR inhibits the proliferation of myeloid precursor cells [80]. In vitro studies showed that AhR could influence the monocyte fate to drive the differentiation of DCs over macrophages, suggesting that AhR plays a key role in the macrophage–dendritic cell balance in inflamed tissues [64]. AhR also inhibits human CD34+ hematopoietic precursor cells from differentiating into monocytes and Langerhans cells (LCs) [81].
3.2. Regulatory T Lymphocytes
- (1)
- Regulatory CD4+Foxp3+T cells. T-regulatory cells include multiple subsets such as CD4+Foxp3+ (Tregs), CD4+Foxp3−IL-10+(Tr1s) and IL-10+CD8 regulatory cells. Tregs express transcription factor Foxp3 [82,83] and differentiate in the thymus or the periphery [84]. They constitutively express inhibitory molecules such as cytotoxic CTLA-4 [85]. These Tregs play a critical role in suppressing tissue inflammation through the release of cytokines such as TGFβ, IL-10 and IL-35. Gut tissue-resident Tregs can be induced in response to dietary and gut microbiota [86,87]. Studies have shown that AhR is important in Tregs by controlling the production of IL-10 and IL-22 [88,89,90,91,92]. Indeed, the immunosuppressive effect of TCDD, an AhR ligand, is linked to the expansion or induction of Treg cells and promotion of the function of Tregs in mice and in humans [89,90]. Another ligand of AhR (2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE)) also suppresses autoimmunity by inducing Tregs [93]. The activation of AhR with ITE can suppress IBD [94] and ameliorates encephalomyelitis (EAE) symptoms [95]. 4-n-nonylphenol, an agonist for AhR, can induce Tregs [96]. Indole and its derivatives from Trp regulate Tregs through the AhR–ligand–Treg axis, thereby affecting the function of Tregs [97,98]. Notably, AhR expression in the Tregs of the spleen and lymph nodes is very low, whereas AhR is highly expressed in intestinal Tregs [99].
- (2)
- Regulatory type 1 (Tr1) cells. Regulatory type 1 (Tr1) cells are an important subset of CD4+ T cells in the control of excessive inflammatory responses [104]. These cells are characterized by IL-10 expression but not Foxp3. Tr1 cells are prominent in chronic infections and immune manipulations in vivo [105]. They are described with regulating activity due to their tolerance to foreign antigens and their capacity to inhibit the proliferation of lymphocytes [106]. The activation of Trp metabolites receptor AhR supports the differentiation of type 1 regulatory T cells (Tr1) [92] and also promotes the differentiation of CD4+Foxp3+ T cells, which can produce IL-10, and control responder T cells [89]. During Tr1-cell differentiation, AhR physically associates with c-Maf and transactivates the IL-10 and IL-21 promoters [88]. AhR also promotes HIF1-α degradation and takes control of Tr1 cell metabolism [92]. In addition, AhR activation can also initiate the differentiation of mucosal-homing Tim3+Lag3+Tr1 cells [107]. Thus, Trp metabolites promote the differentiation and function of not only Treg cells but also other regulatory T cells, such as Tr1 cells.
3.3. Regulatory B Cells
3.4. IL-22-Producing Cells
3.5. CD4+CD8αα+ Lymphocytes
3.6. Other Immune Cells
4. Trp Metabolites from Gut Microbiota and Immune-Associated Diseases
4.1. Inflammatory Bowel Diseases
4.2. Tumors
4.3. Obesity and Metabolic Syndrome
4.4. Diseases in Nervous System
4.5. Infectious Diseases
4.6. Vascular Inflammation and Cardiovascular Diseases
4.7. Hepatic Fibrosis
5. Potential Application of Gut Microbiota-Derived Trp Metabolites in Immunotherapy
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexeev, E.E.; Lanis, J.M.; Kao, D.J.; Campbell, E.L.; Kelly, C.J.; Battista, K.D.; Gerich, M.E.; Jenkins, B.R.; Walk, S.T.; Kominsky, D.J.; et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol. 2018, 188, 1183–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hubbard, T.D.; Murray, I.A.; Perdew, G.H. Indole and Tryptophan Metabolism: Endogenous and Dietary Routes to Ah Receptor Activation. Drug Metab. Dispos. 2015, 43, 1522–1535. [Google Scholar] [CrossRef] [Green Version]
- Whitehead, T.R.; Price, N.P.; Drake, H.L.; Cotta, M.A. Catabolic pathway for the production of skatole and indoleacetic acid by the acetogen Clostridium drakei, Clostridium scatologenes, and swine manure. Appl. Environ. Microbiol. 2008, 74, 1950–1953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, E.A.; Macfarlane, G.T. Enumeration of human colonic bacteria producing phenolic and indolic compounds: Effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J. Appl. Bacteriol. 1996, 81, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, M.T.; Carlson, J.R. Microbial metabolites of tryptophan in the intestinal tract with special reference to skatole. Am. J. Clin. Nutr. 1979, 32, 173–178. [Google Scholar] [CrossRef]
- Vujkovic-Cvijin, I.; Dunham, R.M.; Iwai, S.; Maher, M.C.; Albright, R.G.; Broadhurst, M.J.; Hernandez, R.D.; Lederman, M.M.; Huang, Y.; Somsouk, M.; et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 2013, 5, 193ra91. [Google Scholar] [CrossRef] [Green Version]
- Bansal, T.; Alaniz, R.C.; Wood, T.K.; Jayaraman, A. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc. Natl. Acad. Sci. USA 2010, 107, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Mawe, G.M.; Hoffman, J.M. Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 473–486. [Google Scholar] [CrossRef] [Green Version]
- Holst, J.J. The physiology of glucagon-like peptide 1. Physiol. Rev. 2007, 87, 1409–1439. [Google Scholar] [CrossRef]
- Hill, D.A.; Artis, D. Intestinal bacteria and the regulation of immune cell homeostasis. Annu. Rev. Immunol. 2010, 28, 623–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinde, R.; Hezaveh, K.; Halaby, M.J.; Kloetgen, A.; Chakravarthy, A.; da Silva Medina, T.; Deol, R.; Manion, K.P.; Baglaenko, Y.; Eldh, M.; et al. Apoptotic cell-induced AhR activity is required for immunological tolerance and suppression of systemic lupus erythematosus in mice and humans. Nat. Immunol. 2018, 19, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Manni, G.; Mondanelli, G.; Scalisi, G.; Pallotta, M.T.; Nardi, D.; Padiglioni, E.; Romani, R.; Talesa, V.N.; Puccetti, P.; Fallarino, F.; et al. Pharmacologic Induction of Endotoxin Tolerance in Dendritic Cells by L-Kynurenine. Front. Immunol. 2020, 11, 292. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, X.; Dong, W.; Fang, Y.; Lv, J.; Zhang, T.; Fiskesund, R.; Xie, J.; Liu, J.; Yin, X.; et al. Tumor-Repopulating Cells Induce PD-1 Expression in CD8+ T Cells by Transferring Kynurenine and AhR Activation. Cancer Cell 2018, 33, 480–494.e7. [Google Scholar] [CrossRef] [Green Version]
- Piper, C.J.M.; Rosser, E.C.; Oleinika, K.; Nistala, K.; Krausgruber, T.; Rendeiro, A.F.; Banos, A.; Drozdov, I.; Villa, M.; Thomson, S.; et al. Aryl Hydrocarbon Receptor Contributes to the Transcriptional Program of IL-10-Producing Regulatory B Cells. Cell Rep. 2019, 29, 1878–1892.e7. [Google Scholar] [CrossRef] [Green Version]
- Cervantes-Barragan, L.; Chai, J.N.; Tianero, M.D.; Di Luccia, B.; Ahern, P.P.; Merriman, J.; Cortez, V.S.; Caparon, M.G.; Donia, M.S.; Gilfillan, S.; et al. Lactobacillus reuteri induces gut intraepithelial CD4+CD8α+ T cells. Science 2017, 357, 806–810. [Google Scholar] [CrossRef] [Green Version]
- Zelante, T.; Iannitti, R.G.; Fallarino, F.; Gargaro, M.; De Luca, A.; Moretti, S.; Bartoli, A.; Romani, L. Tryptophan Feeding of the IDO1-AhR Axis in Host-Microbial Symbiosis. Front. Immunol. 2014, 5, 640. [Google Scholar] [CrossRef] [Green Version]
- Schwarcz, R.; Stone, T.W. The kynurenine pathway and the brain: Challenges, controversies and promises. Neuropharmacology 2017, 112, 237–247. [Google Scholar] [CrossRef] [Green Version]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef]
- Shiomi, Y.; Nishiumi, S.; Ooi, M.; Hatano, N.; Shinohara, M.; Yoshie, T.; Kondo, Y.; Furumatsu, K.; Shiomi, H.; Kutsumi, H.; et al. GCMS-based metabolomic study in mice with colitis induced by dextran sulfate sodium. Inflamm. Bowel Dis. 2011, 17, 2261–2274. [Google Scholar] [CrossRef] [PubMed]
- Botticelli, A.; Cerbelli, B.; Lionetto, L.; Zizzari, I.; Salati, M.; Pisano, A.; Federica, M.; Simmaco, M.; Nuti, M.; Marchetti, P. Can IDO activity predict primary resistance to anti-PD-1 treatment in NSCLC? J. Transl. Med. 2018, 16, 219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roager, H.M.; Licht, T.R. Microbial tryptophan catabolites in health and disease. Nat. Commun. 2018, 9, 3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Xu, K.; Liu, H.; Liu, G.; Bai, M.; Peng, C.; Li, T.; Yin, Y. Impact of the Gut Microbiota on Intestinal Immunity Mediated by Tryptophan Metabolism. Front. Cell. Infect. Microbiol. 2018, 8, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoshnevisan, K.; Chehrehgosha, M.; Conant, M.; Meftah, A.M.; Baharifar, H.; Ejtahed, H.S.; Angoorani, P.; Gholami, M.; Sharifi, F.; Maleki, H.; et al. Interactive relationship between Trp metabolites and gut microbiota: The impact on human pathology of disease. J. Appl. Microbiol. 2022, 132, 4186–4207. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, J. Indole as an intercellular signal in microbial communities. FEMS Microbiol. Rev. 2010, 34, 426–444. [Google Scholar] [CrossRef]
- Dodd, D.; Spitzer, M.H.; Van Treuren, W.; Merrill, B.D.; Hryckowian, A.J.; Higginbottom, S.K.; Le, A.; Cowan, T.M.; Nolan, G.P.; Fischbach, M.A.; et al. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 2017, 551, 648–652. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e6. [Google Scholar] [CrossRef] [Green Version]
- Williams, B.B.; Van Benschoten, A.H.; Cimermancic, P.; Donia, M.S.; Zimmermann, M.; Taketani, M.; Ishihara, A.; Kashyap, P.C.; Fraser, J.S.; Fischbach, M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014, 16, 495–503. [Google Scholar] [CrossRef] [Green Version]
- Honore, A.H.; Aunsbjerg, S.D.; Ebrahimi, P.; Thorsen, M.; Benfeldt, C.; Knochel, S.; Skov, T. Metabolic footprinting for investigation of antifungal properties of Lactobacillus paracasei. Anal. Bioanal. Chem. 2016, 408, 83–96. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Zelante, T.; Iannitti, R.G.; Cunha, C.; De Luca, A.; Giovannini, G.; Pieraccini, G.; Zecchi, R.; D’Angelo, C.; Massi-Benedetti, C.; Fallarino, F.; et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, 372–385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mahler, A.; Balogh, A.; Marko, L.; et al. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 2017, 551, 585–589. [Google Scholar] [CrossRef]
- Smith, E.A.; Macfarlane, G.T. Formation of Phenolic and Indolic Compounds by Anaerobic Bacteria in the Human Large Intestine. Microb. Ecol. 1997, 33, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Koga, J.; Adachi, T.; Hidaka, H. Molecular cloning of the gene for indolepyruvate decarboxylase from Enterobacter cloacae. Mol. Gen. Genet. 1991, 226, 10–16. [Google Scholar] [CrossRef]
- Honeyfield, D.C.; Carlson, J.R. Effect of Indoleacetic Acid and Related Indoles on Lactobacillus sp. Strain 11201 Growth, Indoleacetic Acid Catabolism, and 3-Methylindole Formation. Appl. Environ. Microbiol. 1990, 56, 1373–1377. [Google Scholar] [CrossRef] [Green Version]
- Hamady, Z.Z.R.; Farrar, M.D.; Whitehead, T.R.; Holland, K.T.; Lodge, J.P.A.; Carding, S.R. Identification and use of the putative Bacteroides ovatus xylanase promoter for the inducible production of recombinant human proteins. Microbiology 2008, 154, 3165–3174. [Google Scholar] [CrossRef] [Green Version]
- Cook, R.T.; Schlueter, A.J.; Coleman, R.A.; Tygrett, L.; Ballas, Z.K.; Jerrells, T.R.; Nashelsky, M.B.; Ray, N.B.; Haugen, T.H.; Waldschmidt, T.J. Thymocytes, pre-B cells, and organ changes in a mouse model of chronic ethanol ingestion—Absence of subset-specific glucocorticoid-induced immune cell loss. Alcohol. Clin. Exp. Res. 2007, 31, 1746–1758. [Google Scholar] [CrossRef] [Green Version]
- Colabroy, K.L.; Begley, T.P. Tryptophan catabolism: Identification and characterization of a new degradative pathway. J. Bacteriol. 2005, 187, 7866–7869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, Y.; Muraki, T.; Tokuyama, T.; Iwaki, H.; Tatsuno, M.; Lau, P.C. A novel degradative pathway of 2-nitrobenzoate via 3-hydroxyanthranilate in Pseudomonas fluorescens strain KU-7. FEMS Microbiol. Lett. 2000, 190, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Liu, J.; Zhang, D.; Wang, J.; Tan, Y.; Feng, W.; Peng, C. Ginsenoside Rg1 Alleviates Acute Ulcerative Colitis by Modulating Gut Microbiota and Microbial Tryptophan Metabolism. Front. Immunol. 2022, 13, 817600. [Google Scholar] [CrossRef]
- Yang, X.; Dong, B.; An, L.; Zhang, Q.; Chen, Y.; Wang, H.; Song, Z. Ginsenoside Rb1 ameliorates Glycemic Disorder in Mice with High Fat Diet-Induced Obesity via Regulating Gut Microbiota and Amino Acid Metabolism. Front. Pharmacol. 2021, 12, 756491. [Google Scholar] [CrossRef]
- Borisova, M.A.; Snytnikova, O.A.; Litvinova, E.A.; Achasova, K.M.; Babochkina, T.I.; Pindyurin, A.V.; Tsentalovich, Y.P.; Kozhevnikova, E.N. Fucose Ameliorates Tryptophan Metabolism and Behavioral Abnormalities in a Mouse Model of Chronic Colitis. Nutrients 2020, 12, 445. [Google Scholar] [CrossRef] [Green Version]
- Yan, X.; Yan, J.; Xiang, Q.; Wang, F.; Dai, H.; Huang, K.; Fang, L.; Yao, H.; Wang, L.; Zhang, W. Fructooligosaccharides protect against OVA-induced food allergy in mice by regulating the Th17/Treg cell balance using tryptophan metabolites. Food Funct. 2021, 12, 3191–3205. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liu, D.; Wang, Y.; Liu, L.; Li, J.; Yuan, J.; Jiang, Z.; Jiang, Z.; Hsiao, W.W.; Liu, H.; et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut 2022, 71, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Ren, D.; Zhao, Y.; Liu, L.; Yang, X. Fuzhuan Brick Tea Polysaccharide Improved Ulcerative Colitis in Association with Gut Microbiota-Derived Tryptophan Metabolism. J. Agric. Food Chem. 2021, 69, 8448–8459. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, Z.; Wang, B.; He, X.; Tang, J.; Peng, W.; Zhou, J.; Wang, Y. Flammulina velutipes Polysaccharides Modulate Gut Microbiota and Alleviate Carbon Tetrachloride-Induced Hepatic Oxidative Injury in Mice. Front. Microbiol. 2022, 13, 847653. [Google Scholar] [CrossRef]
- Lin, H.; Meng, L.; Sun, Z.; Sun, S.; Huang, X.; Lin, N.; Zhang, J.; Lu, W.; Yang, Q.; Chi, J.; et al. Yellow Wine Polyphenolic Compound Protects Against Doxorubicin-Induced Cardiotoxicity by Modulating the Composition and Metabolic Function of the Gut Microbiota. Circ. Heart Fail. 2021, 14, e008220. [Google Scholar] [CrossRef]
- Jiao, C.; Zhang, Q.; Yang, M.; Ma, J.; Zhao, X.; Tang, N.; Dai, M.; Li, Q.; Jiang, Z.; Huang, X.; et al. Shenling Baizhu San ameliorates ulcerative colitis by regulating the gut microbiota and its tryptophan metabolites: A complementary medicine to mesalamine. J. Ethnopharmacol. 2022, 291, 115145. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Guo, M.; Feng, J.; Gu, Z.; Zhao, J.; Zhang, H.; Wang, G.; Chen, W. Myristica fragrans Extract Regulates Gut Microbes and Metabolites to Attenuate Hepatic Inflammation and Lipid Metabolism Disorders via the AhR-FAS and NF-kappaB Signaling Pathways in Mice with Non-Alcoholic Fatty Liver Disease. Nutrients 2022, 14, 1699. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Luo, L.; Bian, X.; Liu, R.H.; Zhao, S.; Chen, Y.; Sun, K.; Jiang, J.; Liu, Z.; Zeng, L. Pu-erh Tea Restored Circadian Rhythm Disruption by Regulating Tryptophan Metabolism. J. Agric. Food Chem. 2022, 70, 5610–5623. [Google Scholar] [CrossRef]
- Sun, Z.; Li, J.; Wang, W.; Liu, Y.; Liu, J.; Jiang, H.; Lu, Q.; Ding, P.; Shi, R.; Zhao, X.; et al. Qingchang Wenzhong Decoction Accelerates Intestinal Mucosal Healing Through Modulation of Dysregulated Gut Microbiome, Intestinal Barrier and Immune Responses in Mice. Front. Pharmacol. 2021, 12, 738152. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Jian, S.; Wen, C.; Guo, D.; Liao, P.; Wen, J.; Kuang, T.; Han, S.; Liu, Q.; Deng, B. Gallnut Tannic Acid Exerts Anti-stress Effects on Stress-Induced Inflammatory Response, Dysbiotic Gut Microbiota, and Alterations of Serum Metabolic Profile in Beagle Dogs. Front. Nutr. 2022, 9, 847966. [Google Scholar] [CrossRef]
- Wan, Y.; Yang, L.; Li, H.; Ren, H.; Zhu, K.; Dong, Z.; Jiang, S.; Shang, E.; Qian, D.; Duan, J. Zingiber officinale and Panax ginseng ameliorate ulcerative colitis in mice via modulating gut microbiota and its metabolites. J. Chromatogr. B 2022, 1203, 123313. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Cheng, L.; Guo, F.; Tao, S.; Zhang, C.; Ma, L.; Fu, P. Fisetin Improves Hyperuricemia-Induced Chronic Kidney Disease via Regulating Gut Microbiota-Mediated Tryptophan Metabolism and Aryl Hydrocarbon Receptor Activation. J. Agric. Food Chem. 2021, 69, 10932–10942. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Jin, Y.; Zhou, W.; Xiao, T.; Wu, Z.; Su, J.; Gao, H.; Shen, P.; Zheng, B.; Luo, Q.; et al. Rifaximin Modulates the Gut Microbiota to Prevent Hepatic Encephalopathy in Liver Cirrhosis without Impacting the Resistome. Front. Cell. Infect. Microbiol. 2021, 11, 761192. [Google Scholar] [CrossRef]
- Hu, W.; Huang, L.; Zhou, Z.; Yin, L.; Tang, J. Diallyl Disulfide (DADS) Ameliorates Intestinal Candida albicans Infection by Modulating the Gut microbiota and Metabolites and Providing Intestinal Protection in Mice. Front. Cell. Infect. Microbiol. 2021, 11, 743454. [Google Scholar] [CrossRef]
- Veldhoen, M.; Hirota, K.; Westendorf, A.M.; Buer, J.; Dumoutier, L.; Renauld, J.C.; Stockinger, B. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature 2008, 453, 106–109. [Google Scholar] [CrossRef]
- Stockinger, B.; Di Meglio, P.; Gialitakis, M.; Duarte, J.H. The aryl hydrocarbon receptor: Multitasking in the immune system. Annu. Rev. Immunol. 2014, 32, 403–432. [Google Scholar] [CrossRef] [PubMed]
- Gasaly, N.; de Vos, P.; Hermoso, M.A. Impact of Bacterial Metabolites on Gut Barrier Function and Host Immunity: A Focus on Bacterial Metabolism and Its Relevance for Intestinal Inflammation. Front. Immunol. 2021, 12, 658354. [Google Scholar] [CrossRef] [PubMed]
- Vyhlidalova, B.; Krasulova, K.; Pecinkova, P.; Marcalikova, A.; Vrzal, R.; Zemankova, L.; Vanco, J.; Travnicek, Z.; Vondracek, J.; Karasova, M.; et al. Gut Microbial Catabolites of Tryptophan Are Ligands and Agonists of the Aryl Hydrocarbon Receptor: A Detailed Characterization. Int. J. Mol. Sci. 2020, 21, 2614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goudot, C.; Coillard, A.; Villani, A.C.; Gueguen, P.; Cros, A.; Sarkizova, S.; Tang-Huau, T.L.; Bohec, M.; Baulande, S.; Hacohen, N.; et al. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity 2017, 47, 582–596.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bessede, A.; Gargaro, M.; Pallotta, M.T.; Matino, D.; Servillo, G.; Brunacci, C.; Bicciato, S.; Mazza, E.M.; Macchiarulo, A.; Vacca, C.; et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature 2014, 511, 184–190. [Google Scholar] [CrossRef] [Green Version]
- Kimura, A.; Naka, T.; Nakahama, T.; Chinen, I.; Masuda, K.; Nohara, K.; Fujii-Kuriyama, Y.; Kishimoto, T. Aryl hydrocarbon receptor in combination with Stat1 regulates LPS-induced inflammatory responses. J. Exp. Med. 2009, 206, 2027–2035. [Google Scholar] [CrossRef] [Green Version]
- Grosskopf, H.; Walter, K.; Karkossa, I.; von Bergen, M.; Schubert, K. Non-Genomic AhR-Signaling Modulates the Immune Response in Endotoxin-Activated Macrophages After Activation by the Environmental Stressor BaP. Front. Immunol. 2021, 12, 620270. [Google Scholar] [CrossRef]
- Masuda, K.; Kimura, A.; Hanieh, H.; Nguyen, N.T.; Nakahama, T.; Chinen, I.; Otoyo, Y.; Murotani, T.; Yamatodani, A.; Kishimoto, T. Aryl hydrocarbon receptor negatively regulates LPS-induced IL-6 production through suppression of histamine production in macrophages. Int. Immunol. 2011, 23, 637–645. [Google Scholar] [CrossRef] [Green Version]
- Walter, K.; Grosskopf, H.; Karkossa, I.; von Bergen, M.; Schubert, K. Proteomic Characterization of the Cellular Effects of AhR Activation by Microbial Tryptophan Catabolites in Endotoxin-Activated Human Macrophages. Int. J. Environ. Res. Public Health 2021, 18, 10336. [Google Scholar] [CrossRef]
- Zhu, J.; Luo, L.; Tian, L.; Yin, S.; Ma, X.; Cheng, S.; Tang, W.; Yu, J.; Ma, W.; Zhou, X.; et al. Aryl Hydrocarbon Receptor Promotes IL-10 Expression in Inflammatory Macrophages Through Src-STAT3 Signaling Pathway. Front. Immunol. 2018, 9, 2033. [Google Scholar] [CrossRef] [Green Version]
- Campesato, L.F.; Budhu, S.; Tchaicha, J.; Weng, C.H.; Gigoux, M.; Cohen, I.J.; Redmond, D.; Mangarin, L.; Pourpe, S.; Liu, C.; et al. Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine. Nat. Commun. 2020, 11, 4011. [Google Scholar] [CrossRef] [PubMed]
- Opitz, C.A.; Litzenburger, U.M.; Sahm, F.; Ott, M.; Tritschler, I.; Trump, S.; Schumacher, T.; Jestaedt, L.; Schrenk, D.; Weller, M.; et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature 2011, 478, 197–203. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Kwak, J.H.; Pyo, S. Inhibition of LPS-induced inflammatory mediators by 3-hydroxyanthranilic acid in macrophages through suppression of PI3K/NF-kappaB signaling pathways. Food Funct. 2016, 7, 3073–3082. [Google Scholar] [CrossRef]
- Muller, P.A.; Matheis, F.; Mucida, D. Gut macrophages: Key players in intestinal immunity and tissue physiology. Curr. Opin. Immunol. 2020, 62, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Bain, C.C.; Bravo-Blas, A.; Scott, C.L.; Perdiguero, E.G.; Geissmann, F.; Henri, S.; Malissen, B.; Osborne, L.C.; Artis, D.; Mowat, A.M. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat. Immunol. 2014, 15, 929–937. [Google Scholar] [CrossRef] [Green Version]
- De Schepper, S.; Verheijden, S.; Aguilera-Lizarraga, J.; Viola, M.F.; Boesmans, W.; Stakenborg, N.; Voytyuk, I.; Schmidt, I.; Boeckx, B.; Dierckx de Casterle, I.; et al. Self-Maintaining Gut Macrophages Are Essential for Intestinal Homeostasis. Cell 2018, 175, 400–415.e13. [Google Scholar] [CrossRef] [Green Version]
- Niess, J.H.; Brand, S.; Gu, X.; Landsman, L.; Jung, S.; McCormick, B.A.; Vyas, J.M.; Boes, M.; Ploegh, H.L.; Fox, J.G.; et al. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 2005, 307, 254–258. [Google Scholar] [CrossRef] [Green Version]
- Honda, M.; Surewaard, B.G.J.; Watanabe, M.; Hedrick, C.C.; Lee, W.Y.; Brown, K.; McCoy, K.D.; Kubes, P. Perivascular localization of macrophages in the intestinal mucosa is regulated by Nr4a1 and the microbiome. Nat. Commun. 2020, 11, 1329. [Google Scholar] [CrossRef]
- Hadis, U.; Wahl, B.; Schulz, O.; Hardtke-Wolenski, M.; Schippers, A.; Wagner, N.; Muller, W.; Sparwasser, T.; Forster, R.; Pabst, O. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity 2011, 34, 237–246. [Google Scholar] [CrossRef]
- Riemschneider, S.; Kohlschmidt, J.; Fueldner, C.; Esser, C.; Hauschildt, S.; Lehmann, J. Aryl hydrocarbon receptor activation by benzo(a)pyrene inhibits proliferation of myeloid precursor cells and alters the differentiation state as well as the functional phenotype of murine bone marrow-derived macrophages. Toxicol. Lett. 2018, 296, 106–113. [Google Scholar] [CrossRef]
- Platzer, B.; Richter, S.; Kneidinger, D.; Waltenberger, D.; Woisetschlager, M.; Strobl, H. Aryl hydrocarbon receptor activation inhibits in vitro differentiation of human monocytes and Langerhans dendritic cells. J. Immunol. 2009, 183, 66–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khattri, R.; Cox, T.; Yasayko, S.A.; Ramsdell, F. An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat. Immunol. 2003, 4, 337–342. [Google Scholar] [CrossRef]
- Darrigues, J.; van Meerwijk, J.P.M.; Romagnoli, P. Age-Dependent Changes in Regulatory T Lymphocyte Development and Function: A Mini-Review. Gerontology 2018, 64, 28–35. [Google Scholar] [CrossRef] [Green Version]
- Langhans, B.; Nischalke, H.D.; Kramer, B.; Dold, L.; Lutz, P.; Mohr, R.; Vogt, A.; Toma, M.; Eis-Hubinger, A.M.; Nattermann, J.; et al. Role of regulatory T cells and checkpoint inhibition in hepatocellular carcinoma. Cancer Immunol. Immunother. 2019, 68, 2055–2066. [Google Scholar] [CrossRef]
- Russler-Germain, E.V.; Rengarajan, S.; Hsieh, C.S. Antigen-specific regulatory T-cell responses to intestinal microbiota. Mucosal Immunol. 2017, 10, 1375–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.S.; Hong, S.W.; Han, D.; Yi, J.; Jung, J.; Yang, B.G.; Lee, J.Y.; Lee, M.; Surh, C.D. Dietary antigens limit mucosal immunity by inducing regulatory T cells in the small intestine. Science 2016, 351, 858–863. [Google Scholar] [CrossRef]
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.H.; Weiner, H.L.; Kuchroo, V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010, 11, 854–861. [Google Scholar] [CrossRef] [Green Version]
- Gandhi, R.; Kumar, D.; Burns, E.J.; Nadeau, M.; Dake, B.; Laroni, A.; Kozoriz, D.; Weiner, H.L.; Quintana, F.J. Activation of the aryl hydrocarbon receptor induces human type 1 regulatory T cell-like and Foxp3+ regulatory T cells. Nat. Immunol. 2010, 11, 846–853. [Google Scholar] [CrossRef] [Green Version]
- Quintana, F.J.; Basso, A.S.; Iglesias, A.H.; Korn, T.; Farez, M.F.; Bettelli, E.; Caccamo, M.; Oukka, M.; Weiner, H.L. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008, 453, 65–71. [Google Scholar] [CrossRef]
- Haspel, J.A.; Chettimada, S.; Shaik, R.S.; Chu, J.H.; Raby, B.A.; Cernadas, M.; Carey, V.; Process, V.; Hunninghake, G.M.; Ifedigbo, E.; et al. Circadian rhythm reprogramming during lung inflammation. Nat. Commun. 2014, 5, 4753. [Google Scholar] [CrossRef] [PubMed]
- Mascanfroni, I.D.; Takenaka, M.C.; Yeste, A.; Patel, B.; Wu, Y.; Kenison, J.E.; Siddiqui, S.; Basso, A.S.; Otterbein, L.E.; Pardoll, D.M.; et al. Metabolic control of type 1 regulatory T cell differentiation by AHR and HIF1-α. Nat. Med. 2015, 21, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Quintana, F.J.; Murugaiyan, G.; Farez, M.F.; Mitsdoerffer, M.; Tukpah, A.M.; Burns, E.J.; Weiner, H.L. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. USA 2010, 107, 20768–20773. [Google Scholar] [CrossRef] [Green Version]
- Abron, J.D.; Singh, N.P.; Mishra, M.K.; Price, R.L.; Nagarkatti, M.; Nagarkatti, P.S.; Singh, U.P. An endogenous aryl hydrocarbon receptor ligand, ITE, induces regulatory T cells and ameliorates experimental colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2018, 315, G220–G230. [Google Scholar] [CrossRef]
- Hao, N.; Whitelaw, M.L. The emerging roles of AhR in physiology and immunity. Biochem. Pharmacol. 2013, 86, 561–570. [Google Scholar] [CrossRef] [PubMed]
- Bruhs, A.; Haarmann-Stemmann, T.; Frauenstein, K.; Krutmann, J.; Schwarz, T.; Schwarz, A. Activation of the arylhydrocarbon receptor causes immunosuppression primarily by modulating dendritic cells. J. Investig. Dermatol. 2015, 135, 435–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.; Jiang, Y.; Yang, Y.; Shao, J.; Sun, X.; Chen, J.; Dong, L.; Zhang, J. 3,3′-Diindolylmethane alleviates oxazolone-induced colitis through Th2/Th17 suppression and Treg induction. Mol. Immunol. 2013, 53, 335–344. [Google Scholar] [CrossRef]
- Ehrlich, A.K.; Pennington, J.M.; Wang, X.; Rohlman, D.; Punj, S.; Lohr, C.V.; Newman, M.T.; Kolluri, S.K.; Kerkvliet, N.I. Activation of the Aryl Hydrocarbon Receptor by 10-Cl-BBQ Prevents Insulitis and Effector T Cell Development Independently of Foxp3+ Regulatory T Cells in Nonobese Diabetic Mice. J. Immunol. 2016, 196, 264–273. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Qiu, J.; Bostick, J.W.; Ueda, A.; Schjerven, H.; Li, S.; Jobin, C.; Chen, Z.E.; Zhou, L. The Aryl Hydrocarbon Receptor Preferentially Marks and Promotes Gut Regulatory T Cells. Cell Rep. 2017, 21, 2277–2290. [Google Scholar] [CrossRef] [Green Version]
- Mezrich, J.D.; Fechner, J.H.; Zhang, X.; Johnson, B.P.; Burlingham, W.J.; Bradfield, C.A. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010, 185, 3190–3198. [Google Scholar] [CrossRef] [Green Version]
- Goettel, J.A.; Gandhi, R.; Kenison, J.E.; Yeste, A.; Murugaiyan, G.; Sambanthamoorthy, S.; Griffith, A.E.; Patel, B.; Shouval, D.S.; Weiner, H.L.; et al. AHR Activation Is Protective against Colitis Driven by T Cells in Humanized Mice. Cell Rep. 2016, 17, 1318–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tashita, C.; Hoshi, M.; Hirata, A.; Nakamoto, K.; Ando, T.; Hattori, T.; Yamamoto, Y.; Tezuka, H.; Tomita, H.; Hara, A.; et al. Kynurenine plays an immunosuppressive role in 2,4,6-trinitrobenzene sulfate-induced colitis in mice. World J. Gastroenterol. 2020, 26, 918–932. [Google Scholar] [CrossRef] [PubMed]
- Gargaro, M.; Vacca, C.; Massari, S.; Scalisi, G.; Manni, G.; Mondanelli, G.; Mazza, E.M.C.; Bicciato, S.; Pallotta, M.T.; Orabona, C.; et al. Engagement of Nuclear Coactivator 7 by 3-Hydroxyanthranilic Acid Enhances Activation of Aryl Hydrocarbon Receptor in Immunoregulatory Dendritic Cells. Front. Immunol. 2019, 10, 1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roncarolo, M.G.; Gregori, S.; Battaglia, M.; Bacchetta, R.; Fleischhauer, K.; Levings, M.K. Interleukin-10-secreting type 1 regulatory T cells in rodents and humans. Immunol. Rev. 2006, 212, 28–50. [Google Scholar] [CrossRef]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef] [Green Version]
- Vuddamalay, Y.; Attia, M.; Vicente, R.; Pomie, C.; Enault, G.; Leobon, B.; Joffre, O.; Romagnoli, P.; van Meerwijk, J.P. Mouse and human CD8+ CD28low regulatory T lymphocytes differentiate in the thymus. Immunology 2016, 148, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Ehrlich, A.K.; Pennington, J.M.; Tilton, S.; Wang, X.; Marshall, N.B.; Rohlman, D.; Funatake, C.; Punj, S.; O’Donnell, E.; Yu, Z.; et al. AhR activation increases IL-2 production by alloreactive CD4+ T cells initiating the differentiation of mucosal-homing Tim3+ Lag3+ Tr1 cells. Eur. J. Immunol. 2017, 47, 1989–2001. [Google Scholar] [CrossRef] [Green Version]
- Iperi, C.; Bordron, A.; Dueymes, M.; Pers, J.O.; Jamin, C. Metabolic Program of Regulatory B Lymphocytes and Influence in the Control of Malignant and Autoimmune Situations. Front. Immunol. 2021, 12, 735463. [Google Scholar] [CrossRef]
- Shen, P.; Roch, T.; Lampropoulou, V.; O’Connor, R.A.; Stervbo, U.; Hilgenberg, E.; Ries, S.; Dang, V.D.; Jaimes, Y.; Daridon, C.; et al. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 2014, 507, 366–370. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, S.; Manabe, I.; Takaki, S.; Nagasaki, M.; Otsu, M.; Yamashita, H.; Sugita, J.; Yoshimura, K.; Eto, K.; Komuro, I.; et al. Adipose Natural Regulatory B Cells Negatively Control Adipose Tissue Inflammation. Cell Metab. 2013, 18, 759–766. [Google Scholar] [CrossRef] [Green Version]
- Yanaba, K.; Bouaziz, J.D.; Haas, K.M.; Poe, J.C.; Fujimoto, M.; Tedder, T.F. A regulatory B cell subset with a unique CD1dhiCD5+ phenotype controls T cell-dependent inflammatory responses. Immunity 2008, 28, 639–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Zhang, M.; Qi, H.; Gao, Y.; Yang, Y.; Yun, H.; Zhang, Q.; Yang, X.; Zhang, Y.; He, J.; et al. Gut microbiota-derived metabolite 3-idoleacetic acid together with LPS induces IL-35+ B cell generation. Microbiome 2022, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Rosser, E.C.; Mauri, C. The emerging field of regulatory B cell immunometabolism. Cell Metab. 2021, 33, 1088–1097. [Google Scholar] [CrossRef] [PubMed]
- Mauri, C.; Bosma, A. Immune regulatory function of B cells. Annu. Rev. Immunol. 2012, 30, 221–241. [Google Scholar] [CrossRef]
- Wang, R.X.; Yu, C.R.; Dambuza, I.M.; Mahdi, R.M.; Dolinska, M.B.; Sergeev, Y.V.; Wingfield, P.T.; Kim, S.H.; Egwuagu, C.E. Interleukin-35 induces regulatory B cells that suppress autoimmune disease. Nat. Med. 2014, 20, 633–641. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, Y.; Li, C.; Zhang, X.; Cheng, L.; Dai, L.; Wang, Y.; Wang, F.; Shi, G.; Li, Y.; et al. IL-35 Decelerates the Inflammatory Process by Regulating Inflammatory Cytokine Secretion and M1/M2 Macrophage Ratio in Psoriasis. J. Immunol. 2016, 197, 2131–2144. [Google Scholar] [CrossRef]
- Sawant, D.V.; Yano, H.; Chikina, M.; Zhang, Q.; Liao, M.; Liu, C.; Callahan, D.J.; Sun, Z.; Sun, T.; Tabib, T.; et al. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat. Immunol. 2019, 20, 724–735. [Google Scholar] [CrossRef]
- Chen, C.; Xu, H.; Peng, Y.; Luo, H.; Huang, G.X.; Wu, X.J.; Dai, Y.C.; Luo, H.L.; Zhang, J.A.; Zheng, B.Y.; et al. Elevation in the counts of IL-35-producing B cells infiltrating into lung tissue in mycobacterial infection is associated with the downregulation of Th1/Th17 and upregulation of Foxp3+Treg. Sci. Rep. 2020, 10, 13212. [Google Scholar] [CrossRef]
- Vignali, D.A.; Kuchroo, V.K. IL-12 family cytokines: Immunological playmakers. Nat. Immunol. 2012, 13, 722–728. [Google Scholar] [CrossRef] [Green Version]
- Dambuza, I.M.; He, C.; Choi, J.K.; Yu, C.R.; Wang, R.; Mattapallil, M.J.; Wingfield, P.T.; Caspi, R.R.; Egwuagu, C.E. IL-12p35 induces expansion of IL-10 and IL-35-expressing regulatory B cells and ameliorates autoimmune disease. Nat. Commun. 2017, 8, 719. [Google Scholar] [CrossRef]
- Fagarasan, S.; Muramatsu, M.; Suzuki, K.; Nagaoka, H.; Hiai, H.; Honjo, T. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science 2002, 298, 1424–1427. [Google Scholar] [CrossRef] [PubMed]
- Lundell, A.C.; Bjornsson, V.; Ljung, A.; Ceder, M.; Johansen, S.; Lindhagen, G.; Tornhage, C.J.; Adlerberth, I.; Wold, A.E.; Rudin, A. Infant B cell memory differentiation and early gut bacterial colonization. J. Immunol. 2012, 188, 4315–4322. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhu, S.; Xu, G.; Feng, J.; Han, T.; Zhao, F.; She, Y.L.; Liu, S.; Ye, L.; Zhu, Y. Gene Expression and Antiviral Activity of Interleukin-35 in Response to Influenza A Virus Infection. J. Biol. Chem. 2016, 291, 16863–16876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dudakov, J.A.; Hanash, A.M.; van den Brink, M.R. Interleukin-22: Immunobiology and pathology. Annu. Rev. Immunol. 2015, 33, 747–785. [Google Scholar] [CrossRef] [Green Version]
- Spits, H.; Artis, D.; Colonna, M.; Diefenbach, A.; Di Santo, J.P.; Eberl, G.; Koyasu, S.; Locksley, R.M.; McKenzie, A.N.; Mebius, R.E.; et al. Innate lymphoid cells—A proposal for uniform nomenclature. Nat. Rev. Immunol. 2013, 13, 145–149. [Google Scholar] [CrossRef]
- Li, S.; Bostick, J.W.; Zhou, L. Regulation of Innate Lymphoid Cells by Aryl Hydrocarbon Receptor. Front. Immunol. 2017, 8, 1909. [Google Scholar] [CrossRef] [Green Version]
- Liang, S.C.; Tan, X.Y.; Luxenberg, D.P.; Karim, R.; Dunussi-Joannopoulos, K.; Collins, M.; Fouser, L.A. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J. Exp. Med. 2006, 203, 2271–2279. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez-Vazquez, C.; Quintana, F.J. Regulation of the Immune Response by the Aryl Hydrocarbon Receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Cella, M.; McDonald, K.G.; Garlanda, C.; Kennedy, G.D.; Nukaya, M.; Mantovani, A.; Kopan, R.; Bradfield, C.A.; Newberry, R.D.; et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 2011, 13, 144–151. [Google Scholar] [CrossRef]
- Qiu, J.; Heller, J.J.; Guo, X.; Chen, Z.M.; Fish, K.; Fu, Y.X.; Zhou, L. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 2012, 36, 92–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez de Aguero, M.; Ganal-Vonarburg, S.C.; Fuhrer, T.; Rupp, S.; Uchimura, Y.; Li, H.; Steinert, A.; Heikenwalder, M.; Hapfelmeier, S.; Sauer, U.; et al. The maternal microbiota drives early postnatal innate immune development. Science 2016, 351, 1296–1302. [Google Scholar] [CrossRef] [PubMed]
- Ebihara, T.; Song, C.; Ryu, S.H.; Plougastel-Douglas, B.; Yang, L.; Levanon, D.; Groner, Y.; Bern, M.D.; Stappenbeck, T.S.; Colonna, M.; et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol. 2015, 16, 1124–1133. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Heller, J.J.; Bostick, J.W.; Lee, A.; Schjerven, H.; Kastner, P.; Chan, S.; Chen, Z.E.; Zhou, L. Ikaros Inhibits Group 3 Innate Lymphoid Cell Development and Function by Suppressing the Aryl Hydrocarbon Receptor Pathway. Immunity 2016, 45, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Kiss, E.A.; Vonarbourg, C.; Kopfmann, S.; Hobeika, E.; Finke, D.; Esser, C.; Diefenbach, A. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science 2011, 334, 1561–1565. [Google Scholar] [CrossRef]
- Li, S.; Bostick, J.W.; Ye, J.; Qiu, J.; Zhang, B.; Urban, J.F., Jr.; Avram, D.; Zhou, L. Aryl Hydrocarbon Receptor Signaling Cell Intrinsically Inhibits Intestinal Group 2 Innate Lymphoid Cell Function. Immunity 2018, 49, 915–928.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Pu, A.; Sheng, B.; Zhang, Z.; Li, L.; Liu, Z.; Wang, Q.; Li, X.; Ma, Y.; Yu, M.; et al. Aryl hydrocarbon receptor activation modulates CD8αα+TCRαβ+ IELs and suppression of colitis manifestations in mice. Biomed. Pharmacother. 2017, 87, 127–134. [Google Scholar] [CrossRef]
- Wang, J.; Simonavicius, N.; Wu, X.; Swaminath, G.; Reagan, J.; Tian, H.; Ling, L. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J. Biol. Chem. 2006, 281, 22021–22028. [Google Scholar] [CrossRef] [Green Version]
- Frumento, G.; Rotondo, R.; Tonetti, M.; Damonte, G.; Benatti, U.; Ferrara, G.B. Tryptophan-derived catabolites are responsible for inhibition of T and natural killer cell proliferation induced by indoleamine 2,3-dioxygenase. J. Exp. Med. 2002, 196, 459–468. [Google Scholar] [CrossRef] [Green Version]
- Fallarino, F.; Grohmann, U.; Vacca, C.; Bianchi, R.; Orabona, C.; Spreca, A.; Fioretti, M.C.; Puccetti, P. T cell apoptosis by tryptophan catabolism. Cell Death Differ. 2002, 9, 1069–1077. [Google Scholar] [CrossRef]
- Orabona, C.; Puccetti, P.; Vacca, C.; Bicciato, S.; Luchini, A.; Fallarino, F.; Bianchi, R.; Velardi, E.; Perruccio, K.; Velardi, A.; et al. Toward the identification of a tolerogenic signature in IDO-competent dendritic cells. Blood 2006, 107, 2846–2854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanz, T.V.; Becker, S.; Mohapatra, S.R.; Opitz, C.A.; Wick, W.; Platten, M. Suppression of Th1 differentiation by tryptophan supplementation in vivo. Amino Acids 2017, 49, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Chimerel, C.; Emery, E.; Summers, D.K.; Keyser, U.; Gribble, F.M.; Reimann, F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014, 9, 1202–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, G.; McKernan, D.P.; Gaszner, G.; Quigley, E.M.; Cryan, J.F.; Dinan, T.G. A Distinct Profile of Tryptophan Metabolism along the Kynurenine Pathway Downstream of Toll-Like Receptor Activation in Irritable Bowel Syndrome. Front. Pharmacol. 2012, 3, 90. [Google Scholar] [CrossRef] [Green Version]
- Whitfield-Cargile, C.M.; Cohen, N.D.; Chapkin, R.S.; Weeks, B.R.; Davidson, L.A.; Goldsby, J.S.; Hunt, C.L.; Steinmeyer, S.H.; Menon, R.; Suchodolski, J.S.; et al. The microbiota-derived metabolite indole decreases mucosal inflammation and injury in a murine model of NSAID enteropathy. Gut Microbes 2016, 7, 246–261. [Google Scholar] [CrossRef] [Green Version]
- Sofia, M.A.; Ciorba, M.A.; Meckel, K.; Lim, C.K.; Guillemin, G.J.; Weber, C.R.; Bissonnette, M.; Pekow, J.R. Tryptophan Metabolism through the Kynurenine Pathway is Associated with Endoscopic Inflammation in Ulcerative Colitis. Inflamm. Bowel Dis. 2018, 24, 1471–1480. [Google Scholar] [CrossRef]
- Ramos, F.J.; Kaeberlein, M. Ageing: A healthy diet for stem cells. Nature 2012, 486, 477–478. [Google Scholar] [CrossRef]
- Ala, M. Tryptophan metabolites modulate inflammatory bowel disease and colorectal cancer by affecting immune system. Int. Rev. Immunol. 2022, 41, 326–345. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Platten, M.; Nollen, E.A.A.; Rohrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef]
- Sivaprakasam, S.; Bhutia, Y.D.; Ramachandran, S.; Ganapathy, V. Cell-Surface and Nuclear Receptors in the Colon as Targets for Bacterial Metabolites and Its Relevance to Colon Health. Nutrients 2017, 9, 856. [Google Scholar] [CrossRef] [Green Version]
- Dai, G.; Chen, X.; He, Y. The Gut Microbiota Activates AhR Through the Tryptophan Metabolite Kyn to Mediate Renal Cell Carcinoma Metastasis. Front. Nutr. 2021, 8, 712327. [Google Scholar] [CrossRef] [PubMed]
- Park, E.M.; Chelvanambi, M.; Bhutiani, N.; Kroemer, G.; Zitvogel, L.; Wargo, J.A. Targeting the gut and tumor microbiota in cancer. Nat. Med. 2022, 28, 690–703. [Google Scholar] [CrossRef] [PubMed]
- Trezeguet, V.; Fatrouni, H.; Merched, A.J. Immuno-Metabolic Modulation of Liver Oncogenesis by the Tryptophan Metabolism. Cells 2021, 10, 3469. [Google Scholar] [CrossRef] [PubMed]
- Hezaveh, K.; Shinde, R.S.; Klotgen, A.; Halaby, M.J.; Lamorte, S.; Ciudad, M.T.; Quevedo, R.; Neufeld, L.; Liu, Z.Q.; Jin, R.; et al. Tryptophan-derived microbial metabolites activate the aryl hydrocarbon receptor in tumor-associated macrophages to suppress anti-tumor immunity. Immunity 2022, 55, 324–340.e8. [Google Scholar] [CrossRef]
- Mallmann, N.H.; Lima, E.S.; Lalwani, P. Dysregulation of Tryptophan Catabolism in Metabolic Syndrome. Metab. Syndr. Relat. Disord. 2018, 16, 135–142. [Google Scholar] [CrossRef]
- De Mello, V.D.; Paananen, J.; Lindstrom, J.; Lankinen, M.A.; Shi, L.; Kuusisto, J.; Pihlajamaki, J.; Auriola, S.; Lehtonen, M.; Rolandsson, O.; et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci. Rep. 2017, 7, 46337. [Google Scholar] [CrossRef]
- Qi, Q.; Li, J.; Yu, B.; Moon, J.Y.; Chai, J.C.; Merino, J.; Hu, J.; Ruiz-Canela, M.; Rebholz, C.; Wang, Z.; et al. Host and gut microbial tryptophan metabolism and type 2 diabetes: An integrative analysis of host genetics, diet, gut microbiome and circulating metabolites in cohort studies. Gut 2022, 71, 1095–1105. [Google Scholar] [CrossRef]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.C.; Patel, B.; Yan, R.; Blain, M.; et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat. Med. 2016, 22, 586–597. [Google Scholar] [CrossRef]
- Rosas, H.D.; Doros, G.; Bhasin, S.; Thomas, B.; Gevorkian, S.; Malarick, K.; Matson, W.; Hersch, S.M. A systems-level “misunderstanding”: The plasma metabolome in Huntington’s disease. Ann. Clin. Transl. Neurol. 2015, 2, 756–768. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Reddy, M.C.; Ioerger, T.R.; Rothchild, A.C.; Dartois, V.; Schuster, B.M.; Trauner, A.; Wallis, D.; Galaviz, S.; Huttenhower, C.; et al. Tryptophan biosynthesis protects mycobacteria from CD4 T-cell-mediated killing. Cell 2013, 155, 1296–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiering, C.; Wincent, E.; Metidji, A.; Iseppon, A.; Li, Y.; Potocnik, A.J.; Omenetti, S.; Henderson, C.J.; Wolf, C.R.; Nebert, D.W.; et al. Feedback control of AHR signalling regulates intestinal immunity. Nature 2017, 542, 242–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, X.; Li, H.; Anjum, K.; Zhong, X.; Miao, S.; Zheng, G.; Liu, W.; Li, L. Dual Role of Indoles Derived from Intestinal Microbiota on Human Health. Front. Immunol. 2022, 13, 903526. [Google Scholar] [CrossRef] [PubMed]
- Paeslack, N.; Mimmler, M.; Becker, S.; Gao, Z.; Khuu, M.P.; Mann, A.; Malinarich, F.; Regen, T.; Reinhardt, C. Microbiota-derived tryptophan metabolites in vascular inflammation and cardiovascular disease. Amino Acids 2022. [Google Scholar] [CrossRef] [PubMed]
- Teunis, C.; Nieuwdorp, M.; Hanssen, N. Interactions between Tryptophan Metabolism, the Gut Microbiome and the Immune System as Potential Drivers of Non-Alcoholic Fatty Liver Disease (NAFLD) and Metabolic Diseases. Metabolites 2022, 12, 514. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, R.; Ilha, M.; Vaittinen, M.; Kaminska, D.; Mannisto, V.; Karja, V.; Tuomainen, M.; Hanhineva, K.; Romeo, S.; Pajukanta, P.; et al. Indole-3-Propionic Acid, a Gut-Derived Tryptophan Metabolite, Associates with Hepatic Fibrosis. Nutrients 2021, 13, 3509. [Google Scholar] [CrossRef]
- Gargaro, M.; Manni, G.; Scalisi, G.; Puccetti, P.; Fallarino, F. Tryptophan Metabolites at the Crossroad of Immune-Cell Interaction via the Aryl Hydrocarbon Receptor: Implications for Tumor Immunotherapy. Int. J. Mol. Sci. 2021, 22, 4644. [Google Scholar] [CrossRef]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Zhou, W.; Du, Q.; Yang, M.; Ding, Y.; Hu, R. Blockade of IDO-Kynurenine-AhR Axis Ameliorated Colitis-Associated Colon Cancer via Inhibiting Immune Tolerance. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 1179–1199. [Google Scholar] [CrossRef]
- Triplett, T.A.; Garrison, K.C.; Marshall, N.; Donkor, M.; Blazeck, J.; Lamb, C.; Qerqez, A.; Dekker, J.D.; Tanno, Y.; Lu, W.C.; et al. Reversal of indoleamine 2,3-dioxygenase-mediated cancer immune suppression by systemic kynurenine depletion with a therapeutic enzyme. Nat. Biotechnol. 2018, 36, 758–764. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillere, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Virtue, A.T.; McCright, S.J.; Wright, J.M.; Jimenez, M.T.; Mowel, W.K.; Kotzin, J.J.; Joannas, L.; Basavappa, M.G.; Spencer, S.P.; Clark, M.L.; et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 2019, 11, eaav1892. [Google Scholar] [CrossRef] [PubMed]
- Carrillo-Salinas, F.J.; Anastasiou, M.; Ngwenyama, N.; Kaur, K.; Tai, A.; Smolgovsky, S.A.; Jetton, D.; Aronovitz, M.; Alcaide, P. Gut dysbiosis induced by cardiac pressure overload enhances adverse cardiac remodeling in a T cell-dependent manner. Gut Microbes 2020, 12, 1823801. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, R.; Halmi, D.; Demjan, V.; Berkecz, R.; Pipicz, M.; Csont, T. Kynurenine Pathway Metabolites as Potential Clinical Biomarkers in Coronary Artery Disease. Front. Immunol. 2021, 12, 768560. [Google Scholar] [CrossRef]
- Zhang, Z.; Mu, X.; Cao, Q.; Shi, Y.; Hu, X.; Zheng, H. Honeybee gut Lactobacillus modulates host learning and memory behaviors via regulating tryptophan metabolism. Nat. Commun. 2022, 13, 2037. [Google Scholar] [CrossRef]
- Martin, R.; Langella, P. Emerging Health Concepts in the Probiotics Field: Streamlining the Definitions. Front. Microbiol. 2019, 10, 1047. [Google Scholar] [CrossRef] [Green Version]


| Metabolite | Bacterial Species | References | |
|---|---|---|---|
| Indole | Achromobacter liquefaciens Bacteroides ovatus Bacteroides sp. Clostridium limosum Clostridium sp. Corynebacterium acnes Citrobacter koser Desulfovibrio vulgaris | Enterococcus faecalis Escheichia coli Proteus vulgaris Paracolobactrum coliforme Pseudomonas. aeruginosa Salmonella enterica Vibrio cholerae | [23,26] |
| Indole-3-acid-acetic (IAA) | Bacteroides spp. Bacteroides thetaiotaomicron Bacteroides eggerthii Bacteroides ovatus Bacteroides fragilis Bifidobacterium adolescentis Bifidobacterium longum subsp. longum Bifidobacterium spp. Bifidobacterium pseudolongum Clostridium sporogenes Clostridium bartlettii Clostridium spp. Clostridium difficile | Clostridium lituseburense Clostridium paraputrificum Clostridium perfringens Clostridium putrefaciens Clostridium saccharolyticum Clostridium sticklandii Clostridium subterminale Escherichia coli Eubacterium hallii Eubacterium cylindroides Parabacteroides distasonis Peptostreptococcus asscharolyticus | [5,20,27] |
| Indole3-lactic acid (ILA) | Anaerostipes hadrus Anaerostipes caccae Bacteroides thetaiotaomicron Bacteroides eggerthii Bacteroides ovatus Bacteroides fragilis Bifidobacterium adolescentis Bifidobacterium bifidum Bifidobacterium longum subsp. infantis Bifidobacterium longum subsp. longum Bifidobacterium pseudolongum Bifidobacterium spp. Clostridium bartlettii Clostridium perfringens | Clostridium sporogenes Clostridium saccharolyticum Clostridia Escherichia. coli Eubacterium rectale Eubacterium cylindroides Faecalibacterium prausnitzii Lactobacillus murinus Lactobacillus paracasei Lactobacillus reuteri Megamonas hypermegale Parabacteroides distasonis Peptostreptococcus asscharolyticus | [17,23,28,31] |
| Indole-3-propionic acid (IPA) | Bacteroides Clostridium sporogenes Clostridia Peptostreptococcus spp. Escherichia. coli Lactobacillus Peptostreptococcus russellii | Peptostreptococcus asscharolyticus Peptostreptococcus russellii Peptostreptococcus anaerobius Peptostreptococcus stomatis | [20,23,27,29,32] |
| Indoleacrylic acid (IA) | Clostridium sporogenes Peptostreptococcus spp. Peptostreptococcus. russellii Peptostreptococcus anaerobius Peptostreptococcus stomatis | [27,29,33] | |
| Indole-3-aldehyde (IAld) | Lactobacillus johnsonii Lactobacillus. reuteri Lactobacillus. acidophilus Lactobacillus. murinus Lactobacillus spp. | [17,34,35] | |
| Tryptamine | Bacteroides Clostridium sporogenes Escherichia. coli Firmicutes C. sporogenes Ruminococcus gnavus | [30,36] | |
| Indole-3-acetaldehyde (IAAld) | Escherichia coli | [37] | |
| 3-methylindole (skatole) | Bacteroides spp. Bacteroides thetaiotaomicron Butyrivibrio fibrisolvens Clostridium bartlettii Clostridium spp. Clostridium scatologenes | Clostridium drakei Eubacterium cylindroides Eubacterium rectale Lactobacillus spp. Megamonas hypermegale Parabacteroides distasonis | [5,28,38,39,40] |
| 3-hydroxyanthranilic acid (3-HAA) | Pseudomonas fluorescens strain KU-7 Burkholderia cepacia J2315 | [41,42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, X.; Gao, Y.; Yang, R. Gut Microbiota-Derived Tryptophan Metabolites Maintain Gut and Systemic Homeostasis. Cells 2022, 11, 2296. https://doi.org/10.3390/cells11152296
Su X, Gao Y, Yang R. Gut Microbiota-Derived Tryptophan Metabolites Maintain Gut and Systemic Homeostasis. Cells. 2022; 11(15):2296. https://doi.org/10.3390/cells11152296
Chicago/Turabian StyleSu, Xiaomin, Yunhuan Gao, and Rongcun Yang. 2022. "Gut Microbiota-Derived Tryptophan Metabolites Maintain Gut and Systemic Homeostasis" Cells 11, no. 15: 2296. https://doi.org/10.3390/cells11152296