Breakthroughs and Applications of Organ-on-a-Chip Technology
Abstract
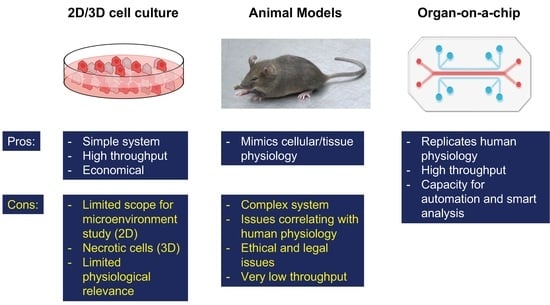
:1. Introduction
2. Types of OOAC Devices
2.1. Lung-on-a-Chip
2.2. Heart-on-a-Chip
2.3. Kidney-on-a-Chip
2.4. Liver-on-a-Chip
2.5. Other OOAC Devices
3. Role of Artificial Intelligence in OOAC
4. OOAC Platforms
5. Applications of OOAC
5.1. Organ/Disease Modelling
5.2. Pharmacology
5.3. Personalized Medicine
5.4. Dentistry
6. Challenges and Future Perspective
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Akhtar, A. The flaws and human harms of animal experimentation. Camb. Q. Healthc. Ethics 2015, 24, 407–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashammakhi, N.A.; Elzagheid, A. Organ-on-a-Chip: New Tool for Personalized Medicine. J. Craniofac. Surg. 2018, 29, 823–824. [Google Scholar] [CrossRef] [PubMed]
- Berndt, E.R.; Nass, D.; Kleinrock, M.; Aitken, M. Decline In Economic Returns From New Drugs Raises Questions About Sustaining Innovations. Health Aff. 2015, 34, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Moon, A. Drug-induced nephrotoxicity and its biomarkers. Biomol. Ther. 2012, 20, 268–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Chen, J.; Bai, H.; Wang, H.; Hao, S.; Ding, Y.; Peng, B.; Zhang, J.; Li, L.; Huang, W. An Overview of Organs-on-Chips Based on Deep Learning. Research 2022, 2022, 9869518. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.B.; Wu, Q.; Yazbeck, J.; Liu, C.; Okhovatian, S.; Radisic, M. Beyond Polydimethylsiloxane: Alternative Materials for Fabrication of Organ-on-a-Chip Devices and Microphysiological Systems. ACS Biomater. Sci. Eng. 2021, 7, 2880–2899. [Google Scholar] [CrossRef]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef] [Green Version]
- Ding, C.; Chen, X.; Kang, Q.; Yan, X. Biomedical Application of Functional Materials in Organ-on-a-Chip. Front. Bioeng. Biotechnol. 2020, 8, 823. [Google Scholar] [CrossRef]
- van Meer, B.J.; de Vries, H.; Firth, K.S.A.; van Weerd, J.; Tertoolen, L.G.J.; Karperien, H.B.J.; Jonkheijm, P.; Denning, C.; Ijerman, A.P.; Mummery, C.L. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem. Biophys. Res. Commun. 2017, 482, 323–328. [Google Scholar] [CrossRef] [Green Version]
- Mujoo, K.; Pandita, R.K.; Tiwari, A.; Charaka, V.; Chakraborty, S.; Singh, D.K.; Hambarde, S.; Hittelman, W.N.; Horikoshi, N.; Hunt, C.R.; et al. Differentiation of Human Induced Pluripotent or Embryonic Stem Cells Decreases the DNA Damage Repair by Homologous Recombination. Stem Cell Rep. 2017, 9, 1660–1674. [Google Scholar] [CrossRef] [Green Version]
- Becker, H.; Hansen-Hagge, T.; Kurtz, A.; Mrowka, R.; Wolfl, S.; Gartner, C. Microfluidic devices for stem-cell cultivation, differentiation and toxicity testing. Proc. Spie Bios 2017, 10061, 1006116. [Google Scholar] [CrossRef]
- Wnorowski, A.; Yang, H.X.; Wu, J.C. Progress, obstacles, and limitations in the use of stem cells in organ-on-a-chip models. Adv. Drug Deliver. Rev. 2019, 140, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.G.; Gupta, A.; Wang, H.; Scherthan, H.; Dhar, S.; Gandhi, V.; Iliakis, G.; Shay, J.W.; Young, C.S.; Pandita, T.K. hTERT associates with human telomeres and enhances genomic stability and DNA repair. Oncogene 2003, 22, 131–146. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Hunt, C.R.; Pandita, R.K.; Kumar, R.; Yang, C.R.; Horikoshi, N.; Bachoo, R.; Serag, S.; Story, M.D.; Shay, J.W.; et al. Lamin A/C depletion enhances DNA damage-induced stalled replication fork arrest. Mol. Cell Biol. 2013, 33, 1210–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Probst, C.; Schneider, S.; Loskill, P. High-throughput organ-on-a-chip systems: Current status and remaining challenges. Curr. Opin. Biomed. Eng. 2018, 6, 33–41. [Google Scholar] [CrossRef]
- Mencattini, A.; Mattei, F.; Schiavoni, G.; Gerardino, A.; Businaro, L.; Di Natale, C.; Martinelli, E. From Petri Dishes to Organ on Chip Platform: The Increasing Importance of Machine Learning and Image Analysis. Front. Pharmacol. 2019, 10, 100. [Google Scholar] [CrossRef] [PubMed]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting organ-level lung functions on a chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Humayun, M.; Chow, C.W.; Young, E.W.K. Microfluidic lung airway-on-a-chip with arrayable suspended gels for studying epithelial and smooth muscle cell interactions. Lab Chip 2018, 18, 1298–1309. [Google Scholar] [CrossRef]
- Park, J.Y.; Ryu, H.; Lee, B.; Ha, D.H.; Ahn, M.; Kim, S.; Kim, J.Y.; Jeon, N.L.; Cho, D.W. Development of a functional airway-on-a-chip by 3D cell printing. Biofabrication 2018, 11, 015002. [Google Scholar] [CrossRef]
- Bai, H.; Si, L.; Jiang, A.; Belgur, C.; Zhai, Y.; Plebani, R.; Oh, C.Y.; Rodas, M.; Patil, A.; Nurani, A.; et al. Mechanical control of innate immune responses against viral infection revealed in a human lung alveolus chip. Nat. Commun. 2022, 13, 1928. [Google Scholar] [CrossRef]
- Mc Namara, K.; Alzubaidi, H.; Jackson, J.K. Cardiovascular disease as a leading cause of death: How are pharmacists getting involved? Integr. Pharm. Res. Pract. 2019, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelleni, M.T. Drug Induced Cardiotoxicity: Mechanism, Prevention and Management; IntechOpen: London, UK, 2018. [Google Scholar]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Ferri, N.; Siegl, P.; Corsini, A.; Herrmann, J.; Lerman, A.; Benghozi, R. Drug attrition during pre-clinical and clinical development: Understanding and managing drug-induced cardiotoxicity. Pharmacol. Ther. 2013, 138, 470–484. [Google Scholar] [CrossRef] [PubMed]
- van Berlo, J.H.; Molkentin, J.D. An emerging consensus on cardiac regeneration. Nat. Med. 2014, 20, 1386–1393. [Google Scholar] [CrossRef] [Green Version]
- Gong, R.; Jiang, Z.; Zagidullin, N.; Liu, T.; Cai, B. Regulation of cardiomyocyte fate plasticity: A key strategy for cardiac regeneration. Signal Transduct. Target. Ther. 2021, 6, 31. [Google Scholar] [CrossRef]
- Mohamed, T.M.A.; Ang, Y.S.; Radzinsky, E.; Zhou, P.; Huang, Y.; Elfenbein, A.; Foley, A.; Magnitsky, S.; Srivastava, D. Regulation of Cell Cycle to Stimulate Adult Cardiomyocyte Proliferation and Cardiac Regeneration. Cell 2018, 173, 104–116.e12. [Google Scholar] [CrossRef] [Green Version]
- Zhu, F.; Meng, Q.; Yu, Y.; Shao, L.; Shen, Z. Adult Cardiomyocyte Proliferation: A New Insight for Myocardial Infarction Therapy. J. Cardiovasc. Transl. Res. 2021, 14, 457–466. [Google Scholar] [CrossRef]
- Jain, A.; Hasan, J.; Desingu, P.A.; Sundaresan, N.R.; Chatterjee, K. Engineering an in vitro organotypic model for studying cardiac hypertrophy. Colloids Surf. B Biointerfaces 2018, 165, 355–362. [Google Scholar] [CrossRef]
- Marsano, A.; Conficconi, C.; Lemme, M.; Occhetta, P.; Gaudiello, E.; Votta, E.; Cerino, G.; Redaelli, A.; Rasponi, M. Beating heart on a chip: A novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 2016, 16, 599–610. [Google Scholar] [CrossRef]
- Agarwal, A.; Goss, J.A.; Cho, A.; McCain, M.L.; Parker, K.K. Microfluidic heart on a chip for higher throughput pharmacological studies. Lab Chip 2013, 13, 3599–3608. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.D.; Liu, L.; Li, J.J.; Yu, L.Q.; Wang, L.; Shi, J.; Chen, Y. Induction and differentiation of human induced pluripotent stem cells into functional cardiomyocytes on a compartmented monolayer of gelatin nanofibers. Nanoscale 2016, 8, 14530–14540. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.S.; Arneri, A.; Bersini, S.; Shin, S.R.; Zhu, K.; Goli-Malekabadi, Z.; Aleman, J.; Colosi, C.; Busignani, F.; Dell’Erba, V.; et al. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016, 110, 45–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Dou, W.; Malhi, M.; Zhu, M.; Liu, H.; Plakhotnik, J.; Xu, Z.; Zhao, Q.; Chen, J.; Chen, S.; et al. Microdevice Platform for Continuous Measurement of Contractility, Beating Rate, and Beating Rhythm of Human-Induced Pluripotent Stem Cell-Cardiomyocytes inside a Controlled Incubator Environment. ACS Appl. Mater. Interfaces 2018, 10, 21173–21183. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; McGuire, A.F.; Lou, H.Y.; Li, T.L.; Tok, J.B.; Cui, B.; Bao, Z. Soft conductive micropillar electrode arrays for biologically relevant electrophysiological recording. Proc. Natl. Acad. Sci. USA 2018, 115, 11718–11723. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Bolonduro, O.A.; Hu, N.; Ju, J.; Rao, A.A.; Duffy, B.M.; Huang, Z.; Black, L.D.; Timko, B.P. Heart-on-a-Chip Model with Integrated Extra- and Intracellular Bioelectronics for Monitoring Cardiac Electrophysiology under Acute Hypoxia. Nano Lett. 2020, 20, 2585–2593. [Google Scholar] [CrossRef]
- Essig, M.; Terzi, F.; Burtin, M.; Friedlander, G. Mechanical strains induced by tubular flow affect the phenotype of proximal tubular cells. Am. J. Physiol. Renal Physiol. 2001, 281, F751–F762. [Google Scholar] [CrossRef]
- Weinberg, E.; Kaazempur-Mofrad, M.; Borenstein, J. Concept and computational design for a bioartificial nephron-on-a-chip. Int. J. Artif. Organs 2008, 31, 508–514. [Google Scholar] [CrossRef]
- Jang, K.J.; Suh, K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip 2010, 10, 36–42. [Google Scholar] [CrossRef]
- Jang, K.J.; Mehr, A.P.; Hamilton, G.A.; McPartlin, L.A.; Chung, S.; Suh, K.Y.; Ingber, D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. 2013, 5, 1119–1129. [Google Scholar] [CrossRef]
- Kim, S.; LesherPerez, S.C.; Kim, B.C.C.; Yamanishi, C.; Labuz, J.M.; Leung, B.; Takayama, S. Pharmacokinetic profile that reduces nephrotoxicity of gentamicin in a perfused kidney-on-a-chip. Biofabrication 2016, 8, 015021. [Google Scholar] [CrossRef]
- Yang, S.H.; Choi, J.W.; Huh, D.; Jo, H.A.; Kim, S.; Lim, C.S.; Lee, J.C.; Kim, H.C.; Kwon, H.M.; Jeong, C.W.; et al. Roles of fluid shear stress and retinoic acid in the differentiation of primary cultured human podocytes. Exp. Cell Res. 2017, 354, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.Y.; Zhang, X.L.; Wen, X.Y.; Wu, T.H.; Wang, W.D.; Yang, M.Z.; Wang, J.; Fang, M.; Lin, B.C.; Lin, H.L. Development of a Functional Glomerulus at the Organ Level on a Chip to Mimic Hypertensive Nephropathy. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, L.N.; Chalasani, N. Epidemiology of Idiosyncratic Drug-Induced Liver Injury. Semin. Liver Dis. 2009, 29, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Kang, Y.B.; Rawat, S.; Duchemin, N.; Bouchard, M.; Noh, M. Human Liver Sinusoid on a Chip for Hepatitis B Virus Replication Study. Micromachines 2017, 8, 27. [Google Scholar] [CrossRef] [Green Version]
- Ortega-Prieto, A.M.; Skelton, J.K.; Wai, S.N.; Large, E.; Lussignol, M.; Vizcay-Barrena, G.; Hughes, D.; Fleck, R.A.; Thursz, M.; Catanese, M.T.; et al. 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection. Nat. Commun. 2018, 9, 682. [Google Scholar] [CrossRef] [Green Version]
- Deng, J.; Chen, Z.Z.; Zhang, X.L.; Luo, Y.; Wu, Z.Z.; Lu, Y.; Liu, T.J.; Zhao, W.J.; Lin, B.C. A liver-chip-based alcoholic liver disease model featuring multi-non-parenchymal cells. Biomed. Microdevices 2019, 21, 57. [Google Scholar] [CrossRef]
- Lou, J.; Zhang, L.; Lv, S.; Zhang, C.; Jiang, S. Biomarkers for Hepatocellular Carcinoma. Biomark. Cancer 2017, 9, 1–9. [Google Scholar] [CrossRef]
- Kane, B.J.; Zinner, M.J.; Yarmush, M.L.; Toner, M. Liver-specific functional studies in a microfluidic array of primary mammalian hepatocytes. Anal. Chem. 2006, 78, 4291–4298. [Google Scholar] [CrossRef]
- Tostoes, R.M.; Leite, S.B.; Serra, M.; Jensen, J.; Bjorquist, P.; Carrondo, M.J.; Brito, C.; Alves, P.M. Human liver cell spheroids in extended perfusion bioreactor culture for repeated-dose drug testing. Hepatology 2012, 55, 1227–1236. [Google Scholar] [CrossRef]
- Ma, L.D.; Wang, Y.T.; Wang, J.R.; Wu, J.L.; Meng, X.S.; Hu, P.; Mu, X.; Liang, Q.L.; Luo, G.A. Design and fabrication of a liver-on-a-chip platform for convenient, highly efficient, and safe in situ perfusion culture of 3D hepatic spheroids. Lab Chip 2018, 18, 2547–2562. [Google Scholar] [CrossRef]
- Allen, J.W.; Hassanein, T.; Bhatia, S.N. Advances in bioartificial liver devices. Hepatology 2001, 34, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Chae, S.; Kim, J.Y.; Han, W.; Kim, J.; Choi, Y.; Cho, D.W. Cell-printed 3D liver-on-a-chip possessing a liver microenvironment and biliary system. Biofabrication 2019, 11, 025001. [Google Scholar] [CrossRef] [PubMed]
- Zamprogno, P.; Wuthrich, S.; Achenbach, S.; Thoma, G.; Stucki, J.D.; Hobi, N.; Schneider-Daum, N.; Lehr, C.M.; Huwer, H.; Geiser, T.; et al. Second-generation lung-on-a-chip with an array of stretchable alveoli made with a biological membrane. Commun. Biol. 2021, 4, 168. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Xiao, Z.; Lv, X.; Zhang, T.; Liu, H. Fabrication and Biomedical Applications of Heart-on-a-chip. Int. J. Bioprint. 2021, 7, 370. [Google Scholar] [CrossRef]
- Kim, S.; Takayama, S. Organ-on-a-chip and the kidney. Kidney Res. Clin. Pract. 2015, 34, 165–169. [Google Scholar] [CrossRef] [Green Version]
- Kanabekova, P.; Kadyrova, A.; Kulsharova, G. Microfluidic Organ-on-a-Chip Devices for Liver Disease Modeling In Vitro. Micromachines 2022, 13, 428. [Google Scholar] [CrossRef]
- Deng, J.; Wei, W.; Chen, Z.; Lin, B.; Zhao, W.; Luo, Y.; Zhang, X. Engineered Liver-on-a-Chip Platform to Mimic Liver Functions and Its Biomedical Applications: A Review. Micromachines 2019, 10, 676. [Google Scholar] [CrossRef] [Green Version]
- Grant, J.; Lee, E.; Almeida, M.; Kim, S.; LoGrande, N.; Goyal, G.; Sesay, A.M.; Breault, D.T.; Prantil-Baun, R.; Ingber, D.E. Establishment of physiologically relevant oxygen gradients in microfluidic organ chips. Lab Chip 2022, 22, 1584–1593. [Google Scholar] [CrossRef]
- Sutterby, E.; Thurgood, P.; Baratchi, S.; Khoshmanesh, K.; Pirogova, E. Microfluidic Skin-on-a-Chip Models: Toward Biomimetic Artificial Skin. Small 2020, 16, e2002515. [Google Scholar] [CrossRef]
- Wufuer, M.; Lee, G.; Hur, W.; Jeon, B.; Kim, B.J.; Choi, T.H.; Lee, S. Skin-on-a-chip model simulating inflammation, edema and drug-based treatment. Sci. Rep. 2016, 6, 37471. [Google Scholar] [CrossRef] [Green Version]
- Lim, H.Y.; Kim, J.; Song, H.J.; Kim, K.; Choi, K.C.; Park, S.; Sung, G.Y. Development of wrinkled skin-on-a-chip (WSOC) by cyclic uniaxial stretching. J. Ind. Eng. Chem. 2018, 68, 238–245. [Google Scholar] [CrossRef]
- O’Neill, A.T.; Monteiro-Riviere, N.A.; Walker, G.M. Characterization of microfluidic human epidermal keratinocyte culture. Cytotechnology 2008, 56, 197–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sriram, G.; Alberti, M.; Dancik, Y.; Wu, B.; Wu, R.G.; Feng, Z.X.; Ramasamy, S.; Bigliardi, P.L.; Bigliardi-Qi, M.; Wang, Z.P. Full-thickness human skin-on-chip with enhanced epidermal morphogenesis and barrier function. Mater. Today 2018, 21, 326–340. [Google Scholar] [CrossRef]
- Sasaki, N.; Tsuchiya, K.; Kobayashi, H. Photolithography-free Skin-on-a-chip for Parallel Permeation Assays. Sensor Mater. 2019, 31, 107–115. [Google Scholar] [CrossRef]
- Mori, N.; Morimoto, Y.; Takeuchi, S. Skin integrated with perfusable vascular channels on a chip. Biomaterials 2017, 116, 48–56. [Google Scholar] [CrossRef]
- Torisawa, Y.S.; Spina, C.S.; Mammoto, T.; Mammoto, A.; Weaver, J.C.; Tat, T.; Collins, J.J.; Ingber, D.E. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat. Methods 2014, 11, 663–669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Q.; Li, X.; Lai, C.; Li, L.; Wu, H.; Wang, Y.; Shi, X. Fabrication of a hydroxyapatite-PDMS microfluidic chip for bone-related cell culture and drug screening. Bioact. Mater. 2021, 6, 169–178. [Google Scholar] [CrossRef]
- Oliver, C.R.; Altemus, M.A.; Westerhof, T.M.; Cheriyan, H.; Cheng, X.; Dziubinski, M.; Wu, Z.F.; Yates, J.; Morikawa, A.; Heth, J.; et al. A platform for artificial intelligence based identification of the extravasation potential of cancer cells into the brain metastatic niche. Lab Chip 2019, 19, 1162–1173. [Google Scholar] [CrossRef]
- Kongadzem, E.-M.L. Machine Learning Application: Organs-on-a-Chip in Parellel; University of Vaasa: Vaasa, Finland, 2018. [Google Scholar]
- Trietsch, S.J.; Naumovska, E.; Kurek, D.; Setyawati, M.C.; Vormann, M.K.; Wilschut, K.J.; Lanz, H.L.; Nicolas, A.; Ng, C.P.; Joore, J.; et al. Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat. Commun. 2017, 8, 262. [Google Scholar] [CrossRef] [Green Version]
- Fetah, K.L.; DiPardo, B.J.; Kongadzem, E.M.; Tomlinson, J.S.; Elzagheid, A.; Elmusrati, M.; Khademhosseini, A.; Ashammakhi, N. Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration. Small 2019, 15, 1901985. [Google Scholar] [CrossRef]
- Ko, J.; Baldassano, S.N.; Loh, P.L.; Kording, K.; Litt, B.; Issadore, D. Machine learning to detect signatures of disease in liquid biopsies—A user’s guide. Lab Chip 2018, 18, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Larsen, B.M.; Kannan, M.; Langer, L.F.; Leibowitz, B.D.; Bentaieb, A.; Cancino, A.; Dolgalev, I.; Drummond, B.E.; Dry, J.R.; Ho, C.S.; et al. A pan-cancer organoid platform for precision medicine. Cell Rep. 2021, 36, 109429. [Google Scholar] [CrossRef]
- Tatara, A.M. Role of Tissue Engineering in COVID-19 and Future Viral Outbreaks. Tissue Eng. Pt. A 2020, 26, 468–474. [Google Scholar] [CrossRef] [Green Version]
- Berkenbrock, J.A.; Grecco-Machado, R.; Achenbach, S. Microfluidic devices for the detection of viruses: Aspects of emergency fabrication during the COVID-19 pandemic and other outbreaks. Proc. R. Soc. 2020, 476, 20200398. [Google Scholar] [CrossRef]
- Gijzen, L.; Yengej, F.A.Y.; Schutgens, F.; Vormann, M.K.; Ammerlaan, C.M.E.; Nicolas, A.; Kurek, D.; Vulto, P.; Rookmaaker, M.B.; Lanz, H.L.; et al. Culture and analysis of kidney tubuloids and perfused tubuloid cells-on-a-chip. Nat. Protoc. 2021, 16, 2023–2050. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.M.; Koo, Y.; Russell, T.; Gay, E.; Li, Y.; Yun, Y. Three-dimensional brain-on-chip model using human iPSC-derived GABAergic neurons and astrocytes: Butyrylcholinesterase post-treatment for acute malathion exposure. PLoS ONE 2020, 15, e0230335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ragelle, H.; Dernick, K.; Khemais, S.; Keppler, C.; Cousin, L.; Farouz, Y.; Louche, C.; Fauser, S.; Kustermann, S.; Tibbitt, M.W.; et al. Human Retinal Microvasculature-on-a-Chip for Drug Discovery. Adv. Healthc. Mater. 2020, 9, e2001531. [Google Scholar] [CrossRef]
- Bircsak, K.M.; DeBiasio, R.; Miedel, M.; Alsebahi, A.; Reddinger, R.; Saleh, A.; Shun, T.Y.; Vernetti, L.A.; Gough, A. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate (R). Toxicology 2021, 450, 152667. [Google Scholar] [CrossRef]
- Eckstrum, K.; Striz, A.; Ferguson, M.; Zhao, Y.; Sprando, R. Evaluation of the utility of the Beta Human Liver Emulation System (BHLES) for CFSAN’s regulatory toxicology program. Food Chem. Toxicol. 2022, 161, 112828. [Google Scholar] [CrossRef]
- Sheyn, D.; Cohn-Yakubovich, D.; Ben-David, S.; De Mel, S.; Chan, V.; Hinojosa, C.; Wen, N.; Hamilton, G.A.; Gazit, D.; Gazit, Z. Bone-chip system to monitor osteogenic differentiation using optical imaging. Microfluid. Nanofluid. 2019, 23, 99. [Google Scholar] [CrossRef]
- Huh, D.; Kim, H.J.; Fraser, J.P.; Shea, D.E.; Khan, M.; Bahinski, A.; Hamilton, G.A.; Ingber, D.E. Microfabrication of human organs-on-chips. Nat. Protoc. 2013, 8, 2135–2157. [Google Scholar] [CrossRef] [PubMed]
- Nawroth, J.C.; Lucchesi, C.; Cheng, D.; Shukla, A.; Ngyuen, J.; Shroff, T.; Varone, A.; Karalis, K.; Lee, H.H.; Alves, S.; et al. A Microengineered Airway Lung Chip Models Key Features of Viral-induced Exacerbation of Asthma. Am. J. Resp. Cell Mol. 2020, 63, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.J.; Otieno, M.A.; Ronxhi, J.; Lim, H.K.; Ewart, L.; Kodella, K.R.; Petropolis, D.B.; Kulkarni, G.; Rubins, J.E.; Conegliano, D.; et al. Reproducing human and cross-species drug toxicities using a Liver-Chip. Sci. Transl. Med. 2019, 11, eaax5516. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.D.; McCoy, L.; Jacobs, E.; Willey, H.; Behn, J.Q.; Nguyen, H.; Bolon, B.; Curley, J.L.; Moore, M.J. Engineering a 3D functional human peripheral nerve in vitro using the Nerve-on-a-Chip platform. Sci. Rep. 2019, 9, 8921. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.L.; Harris, G.; Smirnova, L.; Zufferey, V.; Sa, R.D.D.E.; Russo, F.B.; Braga, P.C.B.B.; Chesnut, M.; Zurich, M.G.; Hogberg, H.T.; et al. Antidepressant Paroxetine Exerts Developmental Neurotoxicity in an iPSC-Derived 3D Human Brain Model. Front. Cell Neurosci. 2020, 14, 25. [Google Scholar] [CrossRef] [Green Version]
- Kundu, A.; McCoy, L.; Azim, N.; Nguyen, H.; Didier, C.M.; Ausaf, T.; Sharma, A.D.; Curley, J.L.; Moore, M.J.; Rajaraman, S. Fabrication and Characterization of 3D Printed, 3D Microelectrode Arrays for Interfacing with a Peripheral Nerve-on-a-Chip. ACS Biomater. Sci. Eng. 2021, 7, 3018–3029. [Google Scholar] [CrossRef]
- Wang, E.Y.; Rafatian, N.; Zhao, Y.; Lee, A.; Lai, B.F.L.; Lu, R.X.; Jekic, D.; Davenport Huyer, L.; Knee-Walden, E.J.; Bhattacharya, S.; et al. Biowire Model of Interstitial and Focal Cardiac Fibrosis. ACS Cent. Sci. 2019, 5, 1146–1158. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Rafatian, N.; Feric, N.T.; Cox, B.J.; Aschar-Sobbi, R.; Wang, E.Y.; Aggarwal, P.; Zhang, B.; Conant, G.; Ronaldson-Bouchard, K.; et al. A Platform for Generation of Chamber-Specific Cardiac Tissues and Disease Modeling. Cell 2019, 176, 913–927.e18. [Google Scholar] [CrossRef] [Green Version]
- Feric, N.T.; Pallotta, I.; Singh, R.; Bogdanowicz, D.R.; Gustilo, M.; Chaudhary, K.; Willette, R.N.; Chendrimada, T.; Xu, X.; Graziano, M.P.; et al. Engineered Cardiac Tissues Generated in the Biowire II: A Platform for Human-Based Drug Discovery. Toxicol. Sci. 2019, 172, 89–97. [Google Scholar] [CrossRef] [Green Version]
- Zamprogno, P.; Thoma, G.; Cencen, V.; Ferrari, D.; Putz, B.; Michler, J.; Fantner, G.E.; Guenat, O.T. Mechanical Properties of Soft Biological Membranes for Organ-on-a-Chip Assessed by Bulge Test and AFM. ACS Biomater. Sci. Eng. 2021, 7, 2990–2997. [Google Scholar] [CrossRef]
- Kiener, M.; De Maddalena, L.; Roldan, N.; Thalmann, G.N.; Geiser, T.; Hobi, N.; Kruithof-De Julio, M. Late Breaking Abstract—Organoids and lung-on-chip to study SARS-CoV-2-mediated lung injury. Eur. Respir. J. 2020, 56, 4149. [Google Scholar] [CrossRef]
- Richter, C.; Hidalgo, A.; Carius, P.; Roldan, N.; Stucki, J.; Hobi, N.; Schneider-Daum, N.; Lehr, C.M. Modelling alveoli on a breathing lung-on-chip in health and disease. Eur. Respir. J. 2020, 56, 3345. [Google Scholar] [CrossRef]
- Materne, E.M.; Maschmeyer, I.; Lorenz, A.K.; Horland, R.; Schimek, K.M.; Busek, M.; Sonntag, F.; Lauster, R.; Marx, U. The multi-organ chip--a microfluidic platform for long-term multi-tissue coculture. J. Vis. Exp. 2015, 98, e52526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoon, J.; Hesse, B.; Rakow, A.; Ort, M.J.; Lagrange, A.; Jacobi, D.; Winter, A.; Huesker, K.; Reinke, S.; Cotte, M.; et al. Metal-Specific Biomaterial Accumulation in Human Peri-Implant Bone and Bone Marrow. Adv. Sci. 2020, 7, 2000412. [Google Scholar] [CrossRef] [PubMed]
- Baert, Y.; Ruetschle, I.; Cools, W.; Oehme, A.; Lorenz, A.; Marx, U.; Goossens, E.; Maschmeyer, I. A multi-organ-chip co-culture of liver and testis equivalents: A first step toward a systemic male reprotoxicity model. Hum. Reprod. 2020, 35, 1029–1044. [Google Scholar] [CrossRef]
- Kuhnl, J.; Tao, T.P.; Brandmair, K.; Gerlach, S.; Rings, T.; Muller-Vieira, U.; Przibilla, J.; Genies, C.; Jaques-Jamin, C.; Schepky, A.; et al. Characterization of application scenario-dependent pharmacokinetics and pharmacodynamic properties of permethrin and hyperforin in a dynamic skin and liver multi-organ-chip model. Toxicology 2021, 448, 152637. [Google Scholar] [CrossRef]
- Tsamandouras, N.; Chen, W.L.K.; Edington, C.D.; Stokes, C.L.; Griffith, L.G.; Cirit, M. Integrated Gut and Liver Microphysiological Systems for Quantitative In Vitro Pharmacokinetic Studies. AAPS J. 2017, 19, 1499–1512. [Google Scholar] [CrossRef] [Green Version]
- Maass, C.; Dallas, M.; LaBarge, M.E.; Shockley, M.; Valdez, J.; Geishecker, E.; Stokes, C.L.; Griffith, L.G.; Cirit, M. Establishing quasi-steady state operations of microphysiological systems (MPS) using tissue-specific metabolic dependencies. Sci. Rep. 2018, 8, 8015. [Google Scholar] [CrossRef]
- Chandorkar, P.; Posch, W.; Zaderer, V.; Blatzer, M.; Steger, M.; Ammann, C.G.; Binder, U.; Hermann, M.; Hortnagl, P.; Lass-Florl, C.; et al. Fast-track development of an in vitro 3D lung/immune cell model to study Aspergillus infections. Sci. Rep. 2017, 7, 11644. [Google Scholar] [CrossRef] [Green Version]
- Ucciferri, N.; Collnot, E.M.; Gaiser, B.K.; Tirella, A.; Stone, V.; Domenici, C.; Lehr, C.M.; Ahluwalia, A. In vitro toxicological screening of nanoparticles on primary human endothelial cells and the role of flow in modulating cell response. Nanotoxicology 2014, 8, 697–708. [Google Scholar] [CrossRef]
- Pu, Y.; Gingrich, J.; Veiga-Lopez, A. A 3-dimensional microfluidic platform for modeling human extravillous trophoblast invasion and toxicological screening. Lab Chip 2021, 21, 546–557. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Mackay, S.; Gordon, D.M.; Anderson, J.D.; Haithcock, D.W.; Garson, C.J.; Tearney, G.J.; Solomon, G.M.; Pant, K.; Prabhakarpandian, B.; et al. Co-cultured microfluidic model of the airway optimized for microscopy and micro-optical coherence tomography imaging. Biomed. Opt. Express 2019, 10, 5414–5430. [Google Scholar] [CrossRef] [PubMed]
- Sasserath, T.; Rumsey, J.W.; McAleer, C.W.; Bridges, L.R.; Long, C.J.; Elbrecht, D.; Schuler, F.; Roth, A.; Bertinetti-LaPatki, C.; Shuler, M.L.; et al. Differential Monocyte Actuation in a Three-Organ Functional Innate Immune System-on-a-Chip. Adv. Sci. 2020, 7, 2000323. [Google Scholar] [CrossRef] [PubMed]
- Boos, J.A.; Misun, P.M.; Michlmayr, A.; Hierlemann, A.; Frey, O. Microfluidic Multitissue Platform for Advanced Embryotoxicity Testing In Vitro. Adv. Sci. 2019, 6, 1900294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Misun, P.M.; Rothe, J.; Schmid, Y.R.F.; Hierlemann, A.; Frey, O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst. Nanoeng. 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakolish, C.; Chen, Z.; Dalaijamts, C.; Mitra, K.; Liu, Y.; Fulton, T.; Wade, T.L.; Kelly, E.J.; Rusyn, I.; Chiu, W.A. Predicting tubular reabsorption with a human kidney proximal tubule tissue-on-a-chip and physiologically-based modeling. Toxicol. In Vitro 2020, 63, 104752. [Google Scholar] [CrossRef]
- Megaw, R.; Abu-Arafeh, H.; Jungnickel, M.; Mellough, C.; Gurniak, C.; Witke, W.; Zhang, W.; Khanna, H.; Mill, P.; Dhillon, B.; et al. Gelsolin dysfunction causes photoreceptor loss in induced pluripotent cell and animal retinitis pigmentosa models. Nat. Commun. 2017, 8, 271. [Google Scholar] [CrossRef]
- Ohlemacher, S.K.; Sridhar, A.; Xiao, Y.; Hochstetler, A.E.; Sarfarazi, M.; Cummins, T.R.; Meyer, J.S. Stepwise Differentiation of Retinal Ganglion Cells from Human Pluripotent Stem Cells Enables Analysis of Glaucomatous Neurodegeneration. Stem Cells 2016, 34, 1553–1562. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; McCain, M.L.; Yang, L.; He, A.; Pasqualini, F.S.; Agarwal, A.; Yuan, H.; Jiang, D.; Zhang, D.; Zangi, L.; et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014, 20, 616–623. [Google Scholar] [CrossRef]
- Zhang, J.; Tuo, J.; Wang, Z.; Zhu, A.; Machalinska, A.; Long, Q. Pathogenesis of Common Ocular Diseases. J. Ophthalmol. 2015, 2015, 734527. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Liu, W.; Wang, Y.; Wang, J.C.; Tu, Q.; Xu, J.; Liu, R.; Shen, S.F.; Wang, J. Investigation of hypoxia-induced myocardial injury dynamics in a tissue interface mimicking microfluidic device. Anal. Chem. 2013, 85, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Muthard, R.W.; Diamond, S.L. Side view thrombosis microfluidic device with controllable wall shear rate and transthrombus pressure gradient. Lab Chip 2013, 13, 1883–1891. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Tao, T.; Su, W.; Yu, H.; Yu, Y.; Qin, J. A disease model of diabetic nephropathy in a glomerulus-on-a-chip microdevice. Lab Chip 2017, 17, 1749–1760. [Google Scholar] [CrossRef] [PubMed]
- Villenave, R.; Lucchesi, C.; Cheng, D.; Lee, H.; Nguyen, J.; Varone, A.; Karalis, K.; Alves, S.; Salmon, M.; Hamilton, G.A. Severe Asthma-On-Chip: A Novel In Vitro Platform To Model Viral-Induced Exacerbations In Asthma. Am. J. Resp. Crit. Care 2017, 195, A4961. [Google Scholar]
- Marzorati, M.; Vanhoecke, B.; De Ryck, T.; Sadaghian Sadabad, M.; Pinheiro, I.; Possemiers, S.; Van den Abbeele, P.; Derycke, L.; Bracke, M.; Pieters, J.; et al. The HMI module: A new tool to study the Host-Microbiota Interaction in the human gastrointestinal tract in vitro. BMC Microbiol. 2014, 14, 133. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, H.; Deng, P.; Tao, T.; Liu, H.; Wu, S.; Chen, W.; Qin, J. Modeling Human Nonalcoholic Fatty Liver Disease (NAFLD) with an Organoids-on-a-Chip System. ACS Biomater. Sci. Eng. 2020, 6, 5734–5743. [Google Scholar] [CrossRef]
- Bongio, M.; Lopa, S.; Gilardi, M.; Bersini, S.; Moretti, M. A 3D vascularized bone remodeling model combining osteoblasts and osteoclasts in a CaP nanoparticle-enriched matrix. Nanomedicine 2016, 11, 1073–1091. [Google Scholar] [CrossRef]
- Pelkonen, A.; Mzezewa, R.; Sukki, L.; Ryynanen, T.; Kreutzer, J.; Hyvarinen, T.; Vinogradov, A.; Aarnos, L.; Lekkala, J.; Kallio, P.; et al. A modular brain-on-a-chip for modelling epileptic seizures with functionally connected human neuronal networks. Biosens. Bioelectron. 2020, 168, 112553. [Google Scholar] [CrossRef]
- Chakrabarty, S.; Quiros-Solano, W.F.; Kuijten, M.M.P.; Haspels, B.; Mallya, S.; Lo, C.S.Y.; Othman, A.; Silvestri, C.; van de Stolpe, A.; Gaio, N.; et al. A Microfluidic Cancer-on-Chip Platform Predicts Drug Response Using Organotypic Tumor Slice Culture. Cancer Res. 2022, 82, 510–520. [Google Scholar] [CrossRef]
- Matthews, H.; Hanison, J.; Nirmalan, N. “Omics”-Informed Drug and Biomarker Discovery: Opportunities, Challenges and Future Perspectives. Proteomes 2016, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Seo, J.; Byun, W.Y.; Alisafaei, F.; Georgescu, A.; Yi, Y.S.; Massaro-Giordano, M.; Shenoy, V.B.; Lee, V.; Bunya, V.Y.; Huh, D. Multiscale reverse engineering of the human ocular surface. Nat. Med. 2019, 25, 1310–1318. [Google Scholar] [CrossRef] [PubMed]
- Wendelboe, A.M.; Raskob, G.E. Global Burden of Thrombosis Epidemiologic Aspects. Circ. Res. 2016, 118, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Korin, N.; Kanapathipillai, M.; Matthews, B.D.; Crescente, M.; Brill, A.; Mammoto, T.; Ghosh, K.; Jurek, S.; Bencherif, S.A.; Bhatta, D.; et al. Shear-Activated Nanotherapeutics for Drug Targeting to Obstructed Blood Vessels. Science 2012, 337, 738–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namdee, K.; Thompson, A.J.; Charoenphol, P.; Eniola-Adefeso, O. Margination Propensity of Vascular-Targeted Spheres from Blood Flow in a Microfluidic Model of Human Microvessels. Langmuir 2013, 29, 2530–2535. [Google Scholar] [CrossRef] [PubMed]
- Haller, D.G.; Cassidy, J.; Clarke, S.J.; Cunningham, D.; Van Cutsem, E.; Hoff, P.M.; Rothenberg, M.L.; Saltz, L.B.; Schmoll, H.J.; Allegra, C.; et al. Potential regional differences for the tolerability profiles of fluoropyrimidines. J. Clin. Oncol. 2008, 26, 2118–2123. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, A.; Mummery, C.L.; Passier, R.; van der Meer, A.D. Personalised organs-on-chips: Functional testing for precision medicine. Lab Chip 2019, 19, 198–205. [Google Scholar] [CrossRef] [Green Version]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.H.; Alves, S.E.; Salmon, M.; Ferrante, T.C.; Weaver, J.C.; et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef]
- Zhang, Y.S.; Aleman, J.; Shin, S.R.; Kilic, T.; Kim, D.; Mousavi Shaegh, S.A.; Massa, S.; Riahi, R.; Chae, S.; Hu, N.; et al. Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors. Proc. Natl. Acad. Sci. USA 2017, 114, E2293–E2302. [Google Scholar] [CrossRef] [Green Version]
- Halaidych, O.V.; Freund, C.; van den Hil, F.; Salvatori, D.C.F.; Riminucci, M.; Mummery, C.L.; Orlova, V.V. Inflammatory Responses and Barrier Function of Endothelial Cells Derived from Human Induced Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 1642–1656. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Franca, C.M.; Tahayeri, A.; Rodrigues, N.S.; Ferdosian, S.; Puppin Rontani, R.M.; Sereda, G.; Ferracane, J.L.; Bertassoni, L.E. The tooth on-a-chip: A microphysiologic model system mimicking the biologic interface of the tooth with biomaterials. Lab Chip 2020, 20, 405–413. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, C.; Rahimi, B.; Padova, D.; Rooholghodos, S.A.; Bienek, D.R.; Luo, X.; Kaufman, G.; Raub, C.B. Oral mucosa-on-a-chip to assess layer-specific responses to bacteria and dental materials. Biomicrofluidics 2018, 12, 054106. [Google Scholar] [CrossRef] [PubMed]
- Niu, L.; Zhang, H.; Liu, Y.; Wang, Y.; Li, A.; Liu, R.; Zou, R.; Yang, Q. Microfluidic Chip for Odontoblasts in Vitro. ACS Biomater. Sci. Eng. 2019, 5, 4844–4851. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Lee, S.S. Shrinkage ratio of PDMS and its alignment method for the wafer level process. Microsyst. Technol. 2008, 14, 205–208. [Google Scholar] [CrossRef]
- Park, D.; Lim, J.; Park, J.Y.; Lee, S.H. Concise Review: Stem Cell Microenvironment on a Chip: Current Technologies for Tissue Engineering and Stem Cell Biology. Stem Cell Transl. Med. 2015, 4, 1352–1368. [Google Scholar] [CrossRef] [Green Version]
- Rusu, E.; Necula, L.G.; Neagu, A.I.; Alecu, M.; Stan, C.; Albulescu, R.; Tanase, C.P. Current status of stem cell therapy: Opportunities and limitations. Turk. J. Biol. 2016, 40, 955–967. [Google Scholar] [CrossRef] [Green Version]
- Wikswo, J.P. Looking to the future of organs-on-chips: Interview with Professor John Wikswo. Future Sci. OA 2017, 3, FSO163. [Google Scholar] [CrossRef] [Green Version]
- Capulli, A.K.; Tian, K.; Mehandru, N.; Bukhta, A.; Choudhury, S.F.; Suchyta, M.; Parker, K.K. Approaching the in vitro clinical trial: Engineering organs on chips. Lab Chip 2014, 14, 3181–3186. [Google Scholar] [CrossRef] [Green Version]
- Kilic, T.; Navaee, F.; Stradolini, F.; Renaud, P.; Carrara, S. Organs-on-chip monitoring: Sensors and other strategies. Microphysiological Syst. 2018, 2, 1–32. [Google Scholar] [CrossRef]
- Ingber, D.E. Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat. Rev. Genet. 2022. [Google Scholar] [CrossRef]
- Fuller, H.C.; Wei, T.Y.; Behrens, M.R.; Ruder, W.C. The Future Application of Organ-on-a-Chip Technologies as Proving Grounds for MicroBioRobots. Micromachines 2020, 11, 947. [Google Scholar] [CrossRef] [PubMed]
- Schuerle, S.; Soleimany, A.P.; Yeh, T.; Anand, G.M.; Haberli, M.; Fleming, H.E.; Mirkhani, N.; Qiu, F.; Hauert, S.; Wang, X.; et al. Synthetic and living micropropellers for convection-enhanced nanoparticle transport. Sci. Adv. 2019, 5, eaav4803. [Google Scholar] [CrossRef] [PubMed]



| Devices | Fabrication Materials | Fabrication Techniques | Features | References |
|---|---|---|---|---|
| Pulmonary edema-on-a-chip | PDMS | Soft lithography |
| [17] |
| Lung airway-on-a-chip | PMMA | Micromilling |
| [18] |
| Airway-on-a-chip | PCL & PDMS | 3D Bioprinting |
| [19] |
| Type of OOAC | Advantages | Disadvantages |
|---|---|---|
| Lung-on-a-chip | Lung-on-a-chip is an excellent replacement to fill the gaps during the transition of test results from an in vitro model to an in vivo environment. One of the pioneering developments in the lung chip platform was the ability to replicate the breathing mechanism [17]. Second-generation lung-on-a-chip studies are aiming to replicate the alveolar network, including physical and biochemical characteristics of alveolar basal membrane by developing stretchable models [54]. | Despite the advancements in lung-on-a-chip, the functional timeline of the chips last up to four weeks only, limiting modelling of chronic conditions. In addition, other characteristics such as cell-to-liquid ratio have to be addressed properly in order to avoid the dilution of metabolites, proteins, and other substances [54]. |
| Heart-on-a-chip | The heart-on-a-chip system helps conduct studies on various cardiac diseases, drug screening and testing. Several associated platforms reviewed in this article have shown high throughput, portability, and the ability to replicate the cardiac system’s physical, electrical, biomechanical characteristics. | Since the heart is a complex structure, it is comparatively difficult to recreate an environment which consist of different types of cells with properties such as polarization and electric impulses to manage contraction of heart chambers, the alignment of these cells, and providing external stimuli. Efforts are being made to overcome this by adopting different fabricating techniques, such as 3D scaffolds and micropattern substrates [55]. |
| Kidney-on-a-chip | The biomimetic kidney-on-a-chip has a significant role in drug toxicological and filtration studies. Various kidney chip models discussed in this article are being improvised at each stage and can retain highly relevant renal characteristics. | Some of the challenges in kidney-on-a-chip include occurrence of bubbles because of smaller dimensions of the chip, degradation of matrix, which can impact cell viability, and optimization of high throughput system. It is highly challenging to maintain consistency in cell seeding as it determines the chip’s characteristics. As mentioned above for other OOAC platforms, the viability and functionality may vary from 3 to 4 weeks [56]. |
| Liver-on-a-chip | It is evident from the various studies that we reviewed on liver-on-a-chip, there is high reproducibility and can highly correlate chemical and toxicological testing. Some of the developed architecture of chip designs can replicate the in vivo physiological environment of the lung more closely. | Despite major advancements, there are still discrepancies in usage of cell sources. For example, stem cell induces hepatocytes and has a stable function, including albumin secretion and urea production. But they require specific induction factors and are costly. Primary hepatocytes can express liver intrinsic characteristics, but are difficult to isolate and incompatible in long term usage. Based on this, biomarker values vary along with discrepancies in the metabolic functions of these cells [57]. Liver chip platforms have low throughput which limits large scale industrial applications [58]. |
| Company | System | Selected Products | Features | Limitations | Region | References |
|---|---|---|---|---|---|---|
| Mimetas |
|
|
|
| The Netherlands | [77,78,79,80] |
| Emulate | Human emulation system to culture multiple organs |
|
|
| USA | [82,83,84,85] |
| AxoSim | Nerve-on-a-chip |
|
|
| USA | [87,88] |
| TARA Biosystems | Heart-on-a-chip |
|
|
| USA | [90,91] |
| AlveoliX | Lung-on-a-chip |
|
|
| Switzerland | [93,94] |
| TissUse | Human-on-a-chip |
|
|
| Germany | [95,96,97,98] |
| CN Bio Innovations |
|
|
|
| UK | [100] |
| Kirkstall |
|
|
|
| UK | [101,102] |
| SynVivo | 3D tissue and OOAC model |
|
|
| USA | [104] |
| Hesperos Inc. |
|
|
|
| USA | [105] |
| InSphero |
|
|
|
| Switzerland | [106,107] |
| Nortis Bio |
|
|
|
| USA | [108] |
| Organs | Devices & Their Purposes | References |
|---|---|---|
| Eye | Age-related macular degeneration model to replicate mechanical stress on retinal pigment epithelial cells | [112] |
| Heart | Heart-on-a-chip platform to analyze hypoxia-induced myocardial injury by using cyanide-p-trifluoromethoxyphenylhydrazone to block oxygen consumption | [113] |
| Vasculature | Microfluidic model to study clot formation useful in the analysis of thrombosis and angiogenesis | [114] |
| Kidney | Model of biomimetic glomerulus-on-a-chip and diabetic kidney to study diabetic nephropathy | [115] |
| Lung | Human airway-on-a-chip was prepared using mucociliary bronchiolar epithelium, which is infected with human rhinovirus to study factors causing asthma | [116] |
| Gut | Human gut-on-a-chip to study gut-immune interactions | [117] |
| Liver | Organoids-on-a-chip using iPSCs to model NAFLD | [118] |
| Bone | In vitro model micro vascularized bone to study the interaction between cell and bone matrix | [119] |
| Brain | Epileptic seizure model of the brain from pluripotent stem cells with the ability to mimic local and circuitry function of brain | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koyilot, M.C.; Natarajan, P.; Hunt, C.R.; Sivarajkumar, S.; Roy, R.; Joglekar, S.; Pandita, S.; Tong, C.W.; Marakkar, S.; Subramanian, L.; et al. Breakthroughs and Applications of Organ-on-a-Chip Technology. Cells 2022, 11, 1828. https://doi.org/10.3390/cells11111828
Koyilot MC, Natarajan P, Hunt CR, Sivarajkumar S, Roy R, Joglekar S, Pandita S, Tong CW, Marakkar S, Subramanian L, et al. Breakthroughs and Applications of Organ-on-a-Chip Technology. Cells. 2022; 11(11):1828. https://doi.org/10.3390/cells11111828
Chicago/Turabian StyleKoyilot, Mufeeda C., Priyadarshini Natarajan, Clayton R. Hunt, Sonish Sivarajkumar, Romy Roy, Shreeram Joglekar, Shruti Pandita, Carl W. Tong, Shamsudheen Marakkar, Lakshminarayanan Subramanian, and et al. 2022. "Breakthroughs and Applications of Organ-on-a-Chip Technology" Cells 11, no. 11: 1828. https://doi.org/10.3390/cells11111828