Endogenous Propionibacterium acnes Promotes Ovarian Cancer Progression via Regulating Hedgehog Signalling Pathway
Abstract
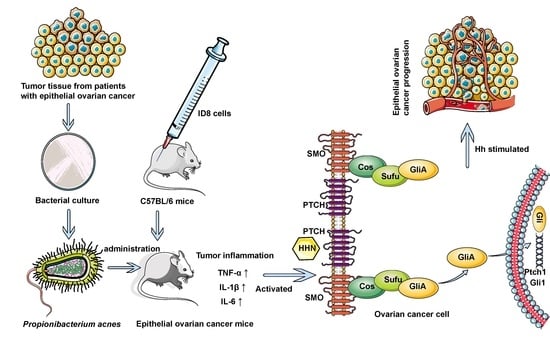
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Enrollment of Patients with EOC and Collection of Tumour Samples
2.2. DNA Extraction and Bacterial 16S rDNA Sequencing
2.3. Bacteria Culture and Identification
2.4. Establishment and Evaluation of Mouse EOC Model
2.5. Hematoxylin-Eosin Staining (HE Staining) and Immunohistochemistry (IHC)
2.6. RNA Extraction and Quantitative PCR
2.7. Western Blotting
2.8. Statistical Analysis
3. Results
3.1. The Taxonomic Diversity of Intratumour Microbiota in EOC and EBOT Tissues
3.2. Isolation of Intratumour Microbiota and Identification of Key Strains
3.3. Intratumour Microbiota in Human Epithelial Ovarian Carcinomas Facilitate EOC Progression in Mice
3.4. Endogenous P. acnes May Promote EOC Progression in Mice via Regulating Hh Signalling Pathway
3.5. Suppression of Hh Signalling Inhibited the EOC Progression Caused by P. acnes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Song, M.; Yeku, O.O.; Rafiq, S.; Purdon, T.; Dong, X.; Zhu, L.; Zhang, T.; Wang, H.; Yu, Z.; Mai, J.; et al. Tumor derived UBR5 promotes ovarian cancer growth and metastasis through inducing immunosuppressive macrophages. Nat. Commun. 2020, 11, 6298. [Google Scholar] [CrossRef] [PubMed]
- Daniels, M.S.; Babb, S.A.; King, R.H.; Urbauer, D.L.; Batte, B.A.; Brandt, A.C.; Amos, C.I.; Buchanan, A.H.; Mutch, D.G.; Lu, K.H. Underestimation of risk of a BRCA1 or BRCA2 mutation in women with high-grade serous ovarian cancer by BRCAPRO: A multi-institution study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 1249–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karakashev, S.; Fukumoto, T.; Zhao, B.; Lin, J.; Wu, S.; Fatkhutdinov, N.; Park, P.H.; Semenova, G.; Jean, S.; Cadungog, M.G.; et al. EZH2 Inhibition Sensitizes CARM1-High, Homologous Recombination Proficient Ovarian Cancers to PARP Inhibition. Cancer Cell 2020, 37, 157–167. [Google Scholar] [CrossRef] [PubMed]
- Krzystyniak, J.; Ceppi, L.; Dizon, D.S.; Birrer, M.J. Epithelial ovarian cancer: The molecular genetics of epithelial ovarian cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2016, 27 (Suppl. S1), i4–i10. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, D.J.; Konner, J.A.; Bell-McGuinn, K.M.; Bhatia, J.; Sabbatini, P.; Aghajanian, C.A.; Offit, K.; Barakat, R.R.; Spriggs, D.R.; Kauff, N.D. Survival in epithelial ovarian cancer: A multivariate analysis incorporating BRCA mutation status and platinum sensitivity. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2011, 22, 1127–1132. [Google Scholar] [CrossRef]
- Vergote, I.B.; Garcia, A.; Micha, J.; Pippitt, C.; Bendell, J.; Spitz, D.; Reed, N.; Dark, G.; Fracasso, P.M.; Ibrahim, E.N.; et al. Randomized multicenter phase II trial comparing two schedules of etirinotecan pegol (NKTR-102) in women with recurrent platinum-resistant/refractory epithelial ovarian cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 4060–4066. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Rustin, G.; Paul, J.; Williams, C.; Pledge, S.; Gabra, H.; Skailes, G.; Lamont, A.; Hindley, A.; Goss, G.; et al. A multicenter, randomized trial of flat dosing versus intrapatient dose escalation of single-agent carboplatin as first-line chemotherapy for advanced ovarian cancer: An SGCTG (SCOTROC 4) and ANZGOG study on behalf of GCIG. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2013, 24, 679–687. [Google Scholar] [CrossRef]
- Mitra, R.; Chen, X.; Greenawalt, E.J.; Maulik, U.; Jiang, W.; Zhao, Z.; Eischen, C.M. Decoding critical long non-coding RNA in ovarian cancer epithelial-to-mesenchymal transition. Nat. Commun. 2017, 8, 1604. [Google Scholar] [CrossRef] [Green Version]
- Uddin, S.; Bu, R.; Ahmed, M.; Abubaker, J.; Al-Dayel, F.; Bavi, P.; Al-Kuraya, K.S. Overexpression of leptin receptor predicts an unfavorable outcome in Middle Eastern ovarian cancer. Mol. Cancer 2009, 8, 74. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Hu, Z.; Shi, H.; Wang, C.; Lei, J.; Cheng, Y. Inhibition of VEGFA Increases the Sensitivity of Ovarian Cancer Cells to Chemotherapy by Suppressing VEGFA-Mediated Autophagy. OncoTargets Ther. 2020, 13, 8161–8171. [Google Scholar] [CrossRef]
- Morita, T.; Mayanagi, T.; Sobue, K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J. Cell Biol. 2007, 179, 1027–1042. [Google Scholar] [CrossRef]
- Louro, I.D.; Bailey, E.C.; Li, X.; South, L.S.; McKie-Bell, P.R.; Yoder, B.K.; Huang, C.C.; Johnson, M.R.; Hill, A.E.; Johnson, R.L.; et al. Comparative gene expression profile analysis of GLI and c-MYC in an epithelial model of malignant transformation. Cancer Res. 2002, 62, 5867–5873. [Google Scholar]
- Zeng, C.; Chen, T.; Zhang, Y.; Chen, Q. Hedgehog signaling pathway regulates ovarian cancer invasion and migration via adhesion molecule CD24. J. Cancer 2017, 8, 786–792. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, Y.; Chen, T.; Zhang, Y.; Xu, R.; Wang, W.; Cheng, M.; Chen, Q. Aberrant Activation Of Hedgehog Signalling Promotes Cell Migration And Invasion Via Matrix Metalloproteinase-7 In Ovarian Cancer Cells. J. Cancer 2019, 10, 990–1003. [Google Scholar] [CrossRef]
- Juarez, V.M.; Montalbine, A.N.; Singh, A. Microbiome as an immune regulator in health, disease, and therapeutics. Adv. Drug Deliv. Rev. 2022, 188, 114400. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef] [Green Version]
- Gérard, P. Gut microbiota and obesity. Cell. Mol. Life Sci. CMLS 2016, 73, 147–162. [Google Scholar] [CrossRef]
- Sun, J.; Kato, I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016, 3, 130–143. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Ren, Z.; Gao, X.; Hu, X.; Zhou, Y.; Jiang, J.; Lu, H.; Yin, S.; Ji, J.; Zhou, L.; et al. Integrated analysis of microbiome and host transcriptome reveals correlations between gut microbiota and clinical outcomes in HBV-related hepatocellular carcinoma. Genome Med. 2020, 12, 102. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef] [PubMed]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e712. [Google Scholar] [CrossRef] [PubMed]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, A.; Yao, B.; Dong, T.; Chen, Y.; Yao, J.; Liu, Y.; Li, H.; Bai, H.; Liu, X.; Zhang, Y.; et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell 2022, 185, 1356–1372. [Google Scholar] [CrossRef]
- Ducie, J.; Dao, F.; Considine, M.; Olvera, N.; Shaw, P.A.; Kurman, R.J.; Shih, I.M.; Soslow, R.A.; Cope, L.; Levine, D.A. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat. Commun. 2017, 8, 990. [Google Scholar] [CrossRef] [Green Version]
- Del Campo, J.M.; Matulonis, U.A.; Malander, S.; Provencher, D.; Mahner, S.; Follana, P.; Waters, J.; Berek, J.S.; Woie, K.; Oza, A.M.; et al. Niraparib Maintenance Therapy in Patients With Recurrent Ovarian Cancer After a Partial Response to the Last Platinum-Based Chemotherapy in the ENGOT-OV16/NOVA Trial. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 2968–2973. [Google Scholar] [CrossRef]
- Banerjee, S.; Wei, Z.; Tan, F.; Peck, K.N.; Shih, N.; Feldman, M.; Rebbeck, T.R.; Alwine, J.C.; Robertson, E.S. Distinct microbiological signatures associated with triple negative breast cancer. Sci. Rep. 2015, 5, 15162. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Tian, T.; Wei, Z.; Shih, N.; Feldman, M.D.; Peck, K.N.; DeMichele, A.M.; Alwine, J.C.; Robertson, E.S. Distinct Microbial Signatures Associated With Different Breast Cancer Types. Front. Microbiol. 2018, 9, 951. [Google Scholar] [CrossRef] [Green Version]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019, 176, 998–1013.e1016. [Google Scholar] [CrossRef] [Green Version]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [PubMed]
- Wegiel, B.; Vuerich, M.; Daneshmandi, S.; Seth, P. Metabolic Switch in the Tumor Microenvironment Determines Immune Responses to Anti-cancer Therapy. Front. Oncol. 2018, 8, 284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota disbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Osman, M.A.; Neoh, H.M.; Ab Mutalib, N.S.; Chin, S.F.; Mazlan, L.; Raja Ali, R.A.; Zakaria, A.D.; Ngiu, C.S.; Ang, M.Y.; Jamal, R. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci. Rep. 2021, 11, 2925. [Google Scholar] [CrossRef] [PubMed]
- Baxter, N.T.; Zackular, J.P.; Chen, G.Y.; Schloss, P.D. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2014, 2, 20. [Google Scholar] [CrossRef] [Green Version]
- Olender, A.; Radej, S.; Płaza, P.; Bar, K.; Maciejewski, R. Propionibacterium acnes infection associated with cancerous prostate hypertrophy. Pol. Arch. Med. Wewn. 2016, 126, 697–699. [Google Scholar] [CrossRef] [Green Version]
- Bae, Y.; Ito, T.; Iida, T.; Uchida, K.; Sekine, M.; Nakajima, Y.; Kumagai, J.; Yokoyama, T.; Kawachi, H.; Akashi, T.; et al. Intracellular Propionibacterium acnes infection in glandular epithelium and stromal macrophages of the prostate with or without cancer. PLoS ONE 2014, 9, e90324. [Google Scholar] [CrossRef]
- Ugge, H.; Udumyan, R.; Carlsson, J.; Andrén, O.; Montgomery, S.; Davidsson, S.; Fall, K. Acne in late adolescence and risk of prostate cancer. Int. J. Cancer 2018, 142, 1580–1585. [Google Scholar] [CrossRef] [Green Version]
- Jia, D.; Nagaoka, Y.; Katsumata, M.; Orsulic, S. Inflammation is a key contributor to ovarian cancer cell seeding. Sci. Rep. 2018, 8, 12394. [Google Scholar] [CrossRef] [Green Version]
- Sapoznik, S.; Bahar-Shany, K.; Brand, H.; Pinto, Y.; Gabay, O.; Glick-Saar, E.; Dor, C.; Zadok, O.; Barshack, I.; Zundelevich, A.; et al. Activation-Induced Cytidine Deaminase Links Ovulation-Induced Inflammation and Serous Carcinogenesis. Neoplasia 2016, 18, 90–99. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.E.; Phan, T.X.; Nguyen, V.H.; Dinh-Vu, H.V.; Zheng, J.H.; Yun, M.; Park, S.G.; Hong, Y.; Choy, H.E.; Szardenings, M.; et al. Salmonella typhimurium Suppresses Tumor Growth via the Pro-Inflammatory Cytokine Interleukin-1β. Theranostics 2015, 5, 1328–1342. [Google Scholar] [CrossRef]
- He, S.; Wang, L.; Miao, L.; Wang, T.; Du, F.; Zhao, L.; Wang, X. Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 2009, 137, 1100–1111. [Google Scholar] [CrossRef] [Green Version]
- Chaudhry, P.; Singh, M.; Triche, T.J.; Guzman, M.; Merchant, A.A. GLI3 repressor determines Hedgehog pathway activation and is required for response to SMO antagonist glasdegib in AML. Blood 2017, 129, 3465–3475. [Google Scholar] [CrossRef]
- Qian, H.; Cao, P.; Hu, M.; Gao, S.; Yan, N.; Gong, X. Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nat. Commun. 2019, 10, 2320. [Google Scholar] [CrossRef] [Green Version]
- Merchant, J.L.; Ding, L. Hedgehog Signaling Links Chronic Inflammation to Gastric Cancer Precursor Lesions. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 201–210. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Q.; Wei, X.; Li, W.; Ma, Y.; Chen, G.; Zhao, L.; Jiang, Y.; Xie, S.; Chen, Q.; Chen, T. Endogenous Propionibacterium acnes Promotes Ovarian Cancer Progression via Regulating Hedgehog Signalling Pathway. Cancers 2022, 14, 5178. https://doi.org/10.3390/cancers14215178
Huang Q, Wei X, Li W, Ma Y, Chen G, Zhao L, Jiang Y, Xie S, Chen Q, Chen T. Endogenous Propionibacterium acnes Promotes Ovarian Cancer Progression via Regulating Hedgehog Signalling Pathway. Cancers. 2022; 14(21):5178. https://doi.org/10.3390/cancers14215178
Chicago/Turabian StyleHuang, Qifa, Xin Wei, Wenyu Li, Yanbing Ma, Guanxiang Chen, Lu Zhao, Ying Jiang, Siqi Xie, Qi Chen, and Tingtao Chen. 2022. "Endogenous Propionibacterium acnes Promotes Ovarian Cancer Progression via Regulating Hedgehog Signalling Pathway" Cancers 14, no. 21: 5178. https://doi.org/10.3390/cancers14215178