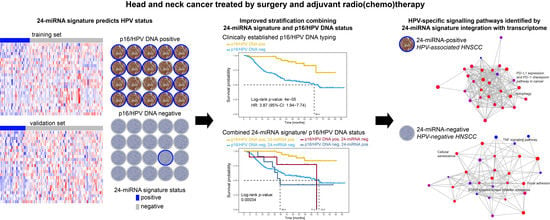
Integration of p16/HPV DNA Status with a 24-miRNA-Defined Molecular Phenotype Improves Clinically Relevant Stratification of Head and Neck Cancer Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohorts
2.2. Tumour Specimens
2.3. HPV Characterisation—LMU-KKG Cohort
2.3.1. p16/HPV DNA Status
2.3.2. HPV E6/E7 RNA Status
2.4. HPV Characterisation—DKTK-ROG Cohort
2.4.1. p16/HPV DNA Status
2.4.2. HPV16 E6/E7 RNA Status
2.5. miRNA Profiling
2.6. Differential miRNA Expression Analysis
2.7. Machine Learning and Performance Testing
2.8. Clinical Endpoints
2.9. Survival Analysis
2.10. Whole RNA Sequencing
2.11. Differential Gene Expression Analysis
2.12. Cancer Signaling Pathway Analysis
2.13. miRNA-mRNA Integration
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, B.; Huang, S.H.; Su, J.; Garden, A.S.; Sturgis, E.M.; Dahlstrom, K.; Lee, N.; Riaz, N.; Pei, X.; Koyfman, S.A.; et al. Development and validation of a staging system for HPV-related oropharyngeal cancer by the International Collaboration on Oropharyngeal cancer Network for Staging (ICON-S): A multicentre cohort study. Lancet Oncol. 2016, 17, 440–451. [Google Scholar] [CrossRef]
- Alsahafi, E.; Begg, K.; Amelio, I.; Raulf, N.; Lucarelli, P.; Sauter, T.; Tavassoli, M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell Death Dis. 2019, 10, 540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, B.; Huang, S.H.; Siu, L.L.; Waldron, J.; Zhao, H.; Perez-Ordonez, B.; Weinreb, I.; Kim, J.; Ringash, J.; Bayley, A.; et al. Deintensification candidate subgroups in human papillomavirus-related oropharyngeal cancer according to minimal risk of distant metastasis. J. Clin. Oncol. 2013, 31, 543–550. [Google Scholar] [CrossRef]
- Hess, A.K.; Muer, A.; Mairinger, F.D.; Weichert, W.; Stenzinger, A.; Hummel, M.; Budach, V.; Tinhofer, I. MiR-200b and miR-155 as predictive biomarkers for the efficacy of chemoradiation in locally advanced head and neck squamous cell carcinoma. Eur. J. Cancer 2017, 77, 3–12. [Google Scholar] [CrossRef]
- Jamali, Z.; Asl Aminabadi, N.; Attaran, R.; Pournagiazar, F.; Ghertasi Oskouei, S.; Ahmadpour, F. MicroRNAs as prognostic molecular signatures in human head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2015, 51, 321–331. [Google Scholar] [CrossRef]
- John, K.; Wu, J.; Lee, B.W.; Farah, C.S. MicroRNAs in Head and Neck Cancer. Int. J. Dent. 2013, 2013, 650218. [Google Scholar] [CrossRef]
- Lajer, C.B.; Garnaes, E.; Friis-Hansen, L.; Norrild, B.; Therkildsen, M.H.; Glud, M.; Rossing, M.; Lajer, H.; Svane, D.; Skotte, L.; et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: Bridging between HPV-related head and neck cancer and cervical cancer. Br. J. Cancer 2012, 106, 1526–1534. [Google Scholar] [CrossRef]
- Sass, S.; Pitea, A.; Unger, K.; Hess, J.; Mueller, N.S.; Theis, F.J. MicroRNA-Target Network Inference and Local Network Enrichment Analysis Identify Two microRNA Clusters with Distinct Functions in Head and Neck Squamous Cell Carcinoma. Int. J. Mol. Sci. 2015, 16, 30204–30222. [Google Scholar] [CrossRef] [Green Version]
- Sethi, N.; Wright, A.; Wood, H.; Rabbitts, P. MicroRNAs and head and neck cancer: Reviewing the first decade of research. Eur. J. Cancer 2014, 50, 2619–2635. [Google Scholar] [CrossRef]
- Hess, J.; Unger, K.; Maihoefer, C.; Schuttrumpf, L.; Wintergerst, L.; Heider, T.; Weber, P.; Marschner, S.; Braselmann, H.; Samaga, D.; et al. A Five-MicroRNA Signature Predicts Survival and Disease Control of Patients with Head and Neck Cancer Negative for HPV Infection. Clin. Cancer Res. 2019, 25, 1505–1516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wald, A.I.; Hoskins, E.E.; Wells, S.I.; Ferris, R.L.; Khan, S.A. Alteration of microRNA profiles in squamous cell carcinoma of the head and neck cell lines by human papillomavirus. Head Neck 2011, 33, 504–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Summerer, I.; Hess, J.; Pitea, A.; Unger, K.; Hieber, L.; Selmansberger, M.; Lauber, K.; Zitzelsberger, H. Integrative analysis of the microRNA-mRNA response to radiochemotherapy in primary head and neck squamous cell carcinoma cells. BMC Genom. 2015, 16, 654. [Google Scholar] [CrossRef] [Green Version]
- McShane, L.M.; Altman, D.G.; Sauerbrei, W.; Taube, S.E.; Gion, M.; Clark, G.M. Statistics Subcommittee of the, N.C.I.E.W.G.o.C.D. REporting recommendations for tumour MARKer prognostic studies (REMARK). Eur. J. Cancer 2005, 41, 1690–1696. [Google Scholar] [CrossRef] [Green Version]
- Simon, R.M.; Paik, S.; Hayes, D.F. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J. Natl. Cancer Inst. 2009, 101, 1446–1452. [Google Scholar] [CrossRef] [Green Version]
- Lohaus, F.; Linge, A.; Tinhofer, I.; Budach, V.; Gkika, E.; Stuschke, M.; Balermpas, P.; Rodel, C.; Avlar, M.; Grosu, A.L.; et al. HPV16 DNA status is a strong prognosticator of loco-regional control after postoperative radiochemotherapy of locally advanced oropharyngeal carcinoma: Results from a multicentre explorative study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG). Radiother. Oncol. 2014, 113, 317–323. [Google Scholar] [CrossRef] [Green Version]
- Maihoefer, C.; Schüttrumpf, L.; Macht, C.; Pflugradt, U.; Hess, J.; Schneider, L.; Woischke, C.; Walch, A.; Baumeister, P.; Kirchner, T.; et al. Postoperative (chemo) radiation in patients with squamous cell cancers of the head and neck-clinical results from the cohort of the clinical cooperation group “Personalized Radiotherapy in Head and Neck Cancer”. Radiat. Oncol. 2018, 13, 123. [Google Scholar] [CrossRef] [Green Version]
- Schuttrumpf, L.; Marschner, S.; Scheu, K.; Hess, J.; Rietzler, S.; Walch, A.; Baumeister, P.; Kirchner, T.; Ganswindt, U.; Zitzelsberger, H.; et al. Definitive chemoradiotherapy in patients with squamous cell cancers of the head and neck-results from an unselected cohort of the clinical cooperation group “Personalized Radiotherapy in Head and Neck Cancer”. Radiat. Oncol. 2020, 15, 7. [Google Scholar] [CrossRef]
- Wintergerst, L.; Selmansberger, M.; Maihoefer, C.; Schuttrumpf, L.; Walch, A.; Wilke, C.; Pitea, A.; Woischke, C.; Baumeister, P.; Kirchner, T.; et al. A prognostic mRNA expression signature of four 16q24.3 genes in radio(chemo)therapy-treated head and neck squamous cell carcinoma (HNSCC). Mol. Oncol. 2018, 12, 2085–2101. [Google Scholar] [CrossRef] [Green Version]
- Ang, K.K.; Harris, J.; Wheeler, R.; Weber, R.; Rosenthal, D.I.; Nguyen-Tan, P.F.; Westra, W.H.; Chung, C.H.; Jordan, R.C.; Lu, C.; et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N. Engl. J. Med. 2010, 363, 24–35. [Google Scholar] [CrossRef] [Green Version]
- Hesselink, A.T.; van den Brule, A.J.; Groothuismink, Z.M.; Molano, M.; Berkhof, J.; Meijer, C.J.; Snijders, P.J. Comparison of three different PCR methods for quantifying human papillomavirus type 16 DNA in cervical scrape specimens. J. Clin. Microbiol. 2005, 43, 4868–4871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindh, M.; Gorander, S.; Andersson, E.; Horal, P.; Mattsby-Balzer, I.; Ryd, W. Real-time Taqman PCR targeting 14 human papilloma virus types. J. Clin. Virol. 2007, 40, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Smeets, S.J.; Hesselink, A.T.; Speel, E.J.; Haesevoets, A.; Snijders, P.J.; Pawlita, M.; Meijer, C.J.; Braakhuis, B.J.; Leemans, C.R.; Brakenhoff, R.H. A novel algorithm for reliable detection of human papillomavirus in paraffin embedded head and neck cancer specimen. Int. J. Cancer 2007, 121, 2465–2472. [Google Scholar] [CrossRef] [PubMed]
- Weber, P.; Kunstner, A.; Hess, J.; Unger, K.; Marschner, S.; Idel, C.; Ribbat-Idel, J.; Baumeister, P.; Gires, O.; Walz, C.; et al. Therapy-Related Transcriptional Subtypes in Matched Primary and Recurrent Head and Neck Cancer. Clin. Cancer Res. 2022, 28, 1038–1052. [Google Scholar] [CrossRef] [PubMed]
- Ndiaye, C.; Mena, M.; Alemany, L.; Arbyn, M.; Castellsague, X.; Laporte, L.; Bosch, F.X.; de Sanjose, S.; Trottier, H. HPV DNA, E6/E7 mRNA, and p16INK4a detection in head and neck cancers: A systematic review and meta-analysis. Lancet Oncol. 2014, 15, 1319–1331. [Google Scholar] [CrossRef]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linge, A.; Lock, S.; Gudziol, V.; Nowak, A.; Lohaus, F.; von Neubeck, C.; Jutz, M.; Abdollahi, A.; Debus, J.; Tinhofer, I.; et al. Low Cancer Stem Cell Marker Expression and Low Hypoxia Identify Good Prognosis Subgroups in HPV(-) HNSCC after Postoperative Radiochemotherapy: A Multicenter Study of the DKTK-ROG. Clin. Cancer Res. 2016, 22, 2639–2649. [Google Scholar] [CrossRef] [Green Version]
- Linge, A.; Schotz, U.; Lock, S.; Lohaus, F.; von Neubeck, C.; Gudziol, V.; Nowak, A.; Tinhofer, I.; Budach, V.; Sak, A.; et al. Comparison of detection methods for HPV status as a prognostic marker for loco-regional control after radiochemotherapy in patients with HNSCC. Radiother. Oncol. 2018, 127, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Schubert, M.; Klinger, B.; Klunemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Bluthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Cava, C.; Colaprico, A.; Bertoli, G.; Graudenzi, A.; Silva, T.C.; Olsen, C.; Noushmehr, H.; Bontempi, G.; Mauri, G.; Castiglioni, I. SpidermiR: An R/Bioconductor Package for Integrative Analysis with miRNA Data. Int. J. Mol. Sci. 2017, 18, 274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samaga, D.; Hornung, R.; Braselmann, H.; Hess, J.; Zitzelsberger, H.; Belka, C.; Boulesteix, A.L.; Unger, K. Single-center versus multi-center data sets for molecular prognostic modeling: A simulation study. Radiat. Oncol. 2020, 15, 109. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, N.C. HPV in non-oropharyngeal head and neck cancer: Does it matter? Ann. Transl. Med. 2020, 8, 1120. [Google Scholar] [CrossRef]
- Watt, K.; Tyryshkin, K.; Renwick, N.; Craig, A.W.B. Distinguishing Tumor and Stromal Sources of MicroRNAs Linked to Metastasis in Cutaneous Melanoma. Transl. Oncol. 2020, 13, 100802. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ma, L.; Jones, T.; Palomero, L.; Pujana, M.A.; Martinez-Ruiz, H.; Ha, P.K.; Murnane, J.; Cuartas, I.; Seoane, J.; et al. Subjugation of TGFbeta Signaling by Human Papilloma Virus in Head and Neck Squamous Cell Carcinoma Shifts DNA Repair from Homologous Recombination to Alternative End Joining. Clin. Cancer Res. 2018, 24, 6001–6014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepore, S.; Lettini, G.; Condelli, V.; Sisinni, L.; Piscazzi, A.; Simeon, V.; Zoppoli, P.; Pedicillo, M.C.; Natalicchio, M.I.; Pietrafesa, M.; et al. Comparative Gene Expression Profiling of Tobacco-Associated HPV-Positive versus Negative Oral Squamous Carcinoma Cell Lines. Int. J. Med. Sci. 2020, 17, 112–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventz, S.; Trippa, L.; Schoenfeld, J.D. Lessons Learned from Deescalation Trials in Favorable Risk HPV-Associated Squamous Cell Head and Neck Cancer-A Perspective on Future Trial Designs. Clin. Cancer Res. 2019, 25, 7281–7286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Zhu, M.; Wang, W.; Chen, D.; Chen, S.; Zheng, H. TNF-α promotes tumor lymph angiogenesis in head and neck squamous cell carcinoma through regulation of ERK3. Transl. Cancer Res. 2019, 8, 2439–2448. [Google Scholar] [CrossRef]
- Hess, J.; Unger, K.; Orth, M.; Schotz, U.; Schuttrumpf, L.; Zangen, V.; Gimenez-Aznar, I.; Michna, A.; Schneider, L.; Stamp, R.; et al. Genomic amplification of Fanconi anemia complementation group A (FancA) in head and neck squamous cell carcinoma (HNSCC): Cellular mechanisms of radioresistance and clinical relevance. Cancer Lett. 2017, 386, 87–99. [Google Scholar] [CrossRef] [PubMed]
- Schoetz, U.; Klein, D.; Hess, J.; Shnayien, S.; Spoerl, S.; Orth, M.; Mutlu, S.; Hennel, R.; Sieber, A.; Ganswindt, U.; et al. Early senescence and production of senescence-associated cytokines are major determinants of radioresistance in head-and-neck squamous cell carcinoma. Cell Death Dis. 2021, 12, 1162. [Google Scholar] [CrossRef] [PubMed]
- Skinner, H.D.; Giri, U.; Yang, L.; Woo, S.H.; Story, M.D.; Pickering, C.R.; Byers, L.A.; Williams, M.D.; El-Naggar, A.; Wang, J.; et al. Proteomic Profiling Identifies PTK2/FAK as a Driver of Radioresistance in HPV-negative Head and Neck Cancer. Clin. Cancer Res. 2016, 22, 4643–4650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyford-Pike, S.; Peng, S.; Young, G.D.; Taube, J.M.; Westra, W.H.; Akpeng, B.; Bruno, T.C.; Richmon, J.D.; Wang, H.; Bishop, J.A.; et al. Evidence for a role of the PD-1:PD-L1 pathway in immune resistance of HPV-associated head and neck squamous cell carcinoma. Cancer Res. 2013, 73, 1733–1741. [Google Scholar] [CrossRef] [Green Version]
- Balermpas, P.; Rodel, F.; Krause, M.; Linge, A.; Lohaus, F.; Baumann, M.; Tinhofer, I.; Budach, V.; Sak, A.; Stuschke, M.; et al. The PD-1/PD-L1 axis and human papilloma virus in patients with head and neck cancer after adjuvant chemoradiotherapy: A multicentre study of the German Cancer Consortium Radiation Oncology Group (DKTK-ROG). Int. J. Cancer 2017, 141, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Grinnell, M.; Krishnan, M.; Ganti, A.K. HPV and the Immune System in Head and Neck Cancers: Therapeutic Considerations. Oncology 2020, 34, 139–143. [Google Scholar]
- Cramer, J.D.; Burtness, B.; Ferris, R.L. Immunotherapy for head and neck cancer: Recent advances and future directions. Oral Oncol. 2019, 99, 104460. [Google Scholar] [CrossRef]





| Parameter | DKTK-ROG | LMU-KKG | ||||||
|---|---|---|---|---|---|---|---|---|
| HPV-Negative (n = 90) | HPV-Positive (n = 38) | Total (n = 128) | p-Value | HPV-Negative (n = 78) | HPV-Positive (n = 23) | Total (n = 101) | p-Value | |
| Age (years) | 0.269 | 0.740 | ||||||
| <45 | 8 (8.9%) | 3 (7.9%) | 11 (8.6%) | 3 (3.8%) | 1 (4.3%) | 4 (4.0%) | ||
| 45–54 | 29 (32.2%) | 8 (21.1%) | 37 (28.9%) | 17 (21.8%) | 6 (26.1%) | 23 (22.8%) | ||
| 55–64 | 36 (40.0%) | 14 (36.8%) | 50 (39.1%) | 28 (35.9%) | 9 (39.1%) | 37 (36.6%) | ||
| 65–74 | 17 (18.9%) | 13 (34.2%) | 30 (23.4%) | 27 (34.6%) | 5 (21.7%) | 32 (31.7%) | ||
| >75 | 3 (3.8%) | 2 (8.7%) | 5 (5.0%) | |||||
| Sex | 0.778 | 0.709 | ||||||
| Male | 73 (81.1%) | 30 (78.9%) | 103 (80.5%) | 51 (65.4%) | 16 (69.6%) | 67 (66.3%) | ||
| Female | 17 (18.9%) | 8 (21.1%) | 25 (19.5%) | 27 (34.6%) | 7 (30.4%) | 34 (33.7%) | ||
| Tumour localization | <0.001 | 0.001 | ||||||
| Hypopharynx | 14 (15.6%) | 1 (2.6%) | 15 (11.7%) | 16 (20.5%) | 1 (4.3%) | 17 (16.8%) | ||
| Oral cavity | 33 (36.7%) | 3 (7.9%) | 36 (28.1%) | 28 (35.9%) | 2 (8.7%) | 30 (29.7%) | ||
| Oropharynx | 43 (47.8%) | 34 (89.5%) | 77 (60.2%) | 34 (43.6%) | 20 (87.0%) | 54 (53.5%) | ||
| UICC TNM stage | 0.221 | 0.773 | ||||||
| I | 2 (2.6%) | 0 (0.0%) | 2 (2.0%) | |||||
| II | 4 (4.4%) | 2 (5.3%) | 6 (4.7%) | 7 (9.0%) | 2 (8.7%) | 9 (8.9%) | ||
| III | 15 (16.7%) | 2 (5.3%) | 17 (13.3%) | 22 (28.2%) | 5 (21.7%) | 27 (26.7%) | ||
| IV | 71 (78.9%) | 34 (89.5%) | 105 (82.0%) | 47 (60.3%) | 16 (69.6%) | 63 (62.4%) | ||
| TNM T stage | 0.601 | 0.834 | ||||||
| T1 | 13 (14.4%) | 4 (10.5%) | 17 (13.3%) | 17 (21.8%) | 5 (21.7%) | 22 (21.8%) | ||
| T2 | 38 (42.2%) | 21 (55.3%) | 59 (46.1%) | 31 (39.7%) | 11 (47.8%) | 42 (41.6%) | ||
| T3 | 22 (24.4%) | 7 (18.4%) | 29 (22.7%) | 18 (23.1%) | 5 (21.7%) | 23 (22.8%) | ||
| T4 | 17 (18.9%) | 6 (15.8%) | 23 (18.0%) | 12 (15.4%) | 2 (8.7%) | 14 (13.9%) | ||
| TNM Nstage | 0.528 | 0.234 | ||||||
| N0 | 12 (13.3%) | 3 (7.9%) | 15 (11.7%) | 20 (25.6%) | 7 (30.4%) | 27 (26.7%) | ||
| N1 | 10 (11.1%) | 4 (10.5%) | 14 (10.9%) | 21 (26.9%) | 2 (8.7%) | 23 (22.8%) | ||
| N2 | 59 (65.6%) | 24 (63.2%) | 83 (64.8%) | 35 (44.9%) | 14 (60.9%) | 49 (48.5%) | ||
| N3 | 9 (10.0%) | 7 (18.4%) | 16 (12.5%) | 2 (2.6%) | 0 (0.0%) | 2 (2.0%) | ||
| Lymphovascular invasion (LVI) | 0.372 | 0.194 | ||||||
| 0 | 46 (63.9%) | 24 (72.7%) | 70 (66.7%) | 52 (77.6%) | 14 (63.6%) | 66 (74.2%) | ||
| 1 | 26 (36.1%) | 9 (27.3%) | 35 (33.3%) | 15 (22.4%) | 8 (36.4%) | 23 (25.8%) | ||
| Missing information | 18 | 5 | 23 | 11 | 1 | 12 | ||
| Venous tumour invasion (VTI) | 0.244 | 0.331 | ||||||
| 0 | 65 (90.3%) | 31 (96.9%) | 96 (92.3%) | 66 (95.7%) | 21 (100.0%) | 87 (96.7%) | ||
| 1 | 7 (9.7%) | 1 (3.1%) | 8 (7.7%) | 3 (4.3%) | 0 (0.0%) | 3 (3.3%) | ||
| Missing information | 18 | 6 | 24 | 9 | 2 | 11 | ||
| Perineural invasion (PNI) | 0.485 | |||||||
| 0 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 37 (74.0%) | 14 (82.4%) | 51 (76.1%) | ||
| 1 | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 13 (26.0%) | 3 (17.6%) | 16 (23.9%) | ||
| Missing information | 90 | 38 | 128 | 28 | 6 | 34 | ||
| Resection margin status | 0.157 | 0.361 | ||||||
| 0 | 47 (52.2%) | 25 (65.8%) | 72 (56.2%) | 59 (77.6%) | 15 (65.2%) | 74 (74.7%) | ||
| 1 | 43 (47.8%) | 13 (34.2%) | 56 (43.8%) | 16 (21.1%) | 8 (34.8%) | 24 (24.2%) | ||
| 2 | 1 (1.3%) | 0 (0.0%) | 1 (1.0%) | |||||
| Missing information | 2 | 0 | 2 | |||||
| ECE | 0.595 | 0.828 | ||||||
| Not applicable (N0) | 12 (13.3%) | 3 (7.9%) | 15 (11.7%) | 20 (26.0%) | 7 (30.4%) | 27 (27.0%) | ||
| No | 33 (36.7%) | 13 (34.2%) | 46 (35.9%) | 32 (41.6%) | 10 (43.5%) | 42 (42.0%) | ||
| Yes | 45 (50.0%) | 22 (57.9%) | 67 (52.3%) | 25 (32.5%) | 6 (26.1%) | 31 (31.0%) | ||
| Missing information | 0 | 0 | 0 | 1 | 0 | 1 | ||
| Smoking status | 0.050 | 0.078 | ||||||
| Missing information | 30 | 13 | 43 | 18 (23.1%) | 5 (21.7%) | 23 (22.8%) | ||
| Nonsmoker | 5 (8.3%) | 6 (24.0%) | 11 (12.9%) | 3 (3.8%) | 4 (17.4%) | 7 (6.9%) | ||
| Smoker | 55 (91.7%) | 19 (76.0%) | 74 (87.1%) | 57 (73.1%) | 14 (60.9%) | 71 (70.3%) | ||
| Simultaneous chemotherapy | 0.553 | |||||||
| Yes | 90 (100.0%) | 38 (100.0%) | 128 (100.0%) | 49 (62.8%) | 16 (69.6%) | 65 (64.4%) | ||
| No | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 29 (37.2%) | 7 (30.4%) | 36 (35.6%) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hess, J.; Unger, K.; Maihoefer, C.; Schüttrumpf, L.; Weber, P.; Marschner, S.; Wintergerst, L.; Pflugradt, U.; Baumeister, P.; Walch, A.; et al. Integration of p16/HPV DNA Status with a 24-miRNA-Defined Molecular Phenotype Improves Clinically Relevant Stratification of Head and Neck Cancer Patients. Cancers 2022, 14, 3745. https://doi.org/10.3390/cancers14153745
Hess J, Unger K, Maihoefer C, Schüttrumpf L, Weber P, Marschner S, Wintergerst L, Pflugradt U, Baumeister P, Walch A, et al. Integration of p16/HPV DNA Status with a 24-miRNA-Defined Molecular Phenotype Improves Clinically Relevant Stratification of Head and Neck Cancer Patients. Cancers. 2022; 14(15):3745. https://doi.org/10.3390/cancers14153745
Chicago/Turabian StyleHess, Julia, Kristian Unger, Cornelius Maihoefer, Lars Schüttrumpf, Peter Weber, Sebastian Marschner, Ludmila Wintergerst, Ulrike Pflugradt, Philipp Baumeister, Axel Walch, and et al. 2022. "Integration of p16/HPV DNA Status with a 24-miRNA-Defined Molecular Phenotype Improves Clinically Relevant Stratification of Head and Neck Cancer Patients" Cancers 14, no. 15: 3745. https://doi.org/10.3390/cancers14153745