Regulation of Long Non-Coding RNAs by Plant Secondary Metabolites: A Novel Anticancer Therapeutic Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methodology for Literature Search and Selection
3. Non-Coding RNAs
3.1. Categories of lncRNAs
3.2. Functions of lncRNAs
4. Role of lncRNAs in Human Cancers
4.1. Role in EMT
4.2. Role in NF-κB Signaling and Telomerase Activity
4.3. Role in Energy Metabolism
4.4. Role in Drug Resistance
5. Plant lncRNAs
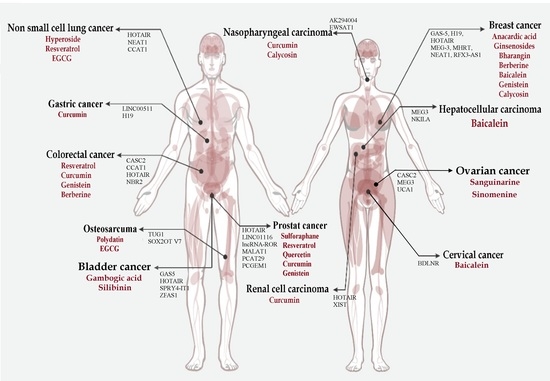
6. Phytochemicals as Antitumor Agents by Targeting lncRNAs
6.1. Anacardic Acids
6.2. Baicalein
6.3. Berberine
6.4. Bharangin
6.5. Calycosin
6.6. Curcumin
6.7. 3,3′-Diindolylmethane (DIM)
6.8. Epigallocatechin-3-Gallate (EGCG)
6.9. Gambogic Acid
6.10. Genistein
6.11. Ginsenosides
6.12. Hyperoside
6.13. Luteolin
6.14. Polydatin
6.15. Quercetin
6.16. Resveratrol
6.17. Sanguinarine
6.18. Silibinin
6.19. Sulforaphane
7. Mechanism of Actions of Phytochemicals in the Regulation of lncRNAs in Cancer
8. Phytochemical-Based Regulation of lncRNAs in Cancer Precision Medicine
9. Conclusions, Current Challenges, and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| BC | bladder cancer |
| BANCR | BRAF-activated long noncoding RNA |
| capn4 | calpain small subunit 1 |
| CCAT1 | colon cancer-associated transcript 1 |
| CTR1 | copper transporter protein 1 |
| DIM | 3,3′-diindolylmethane |
| DNMTs | DNA methyltransferases |
| TOP2A | DNA topoisomerase 2A |
| EGCG | epigallocatechin-3-gallate |
| EMT | epithelial-to-mesenchymal transition |
| EWSAT1 | Ewing sarcoma-associated transcript 1 |
| GA | gambogic acid |
| GAS5 | growth arrest-specific 5 |
| GRB2 | growth factor receptor-bound protein 2 |
| HCC | hepatocellular carcinoma |
| HDACs | histone deacetylases |
| HIF-1α | hypoxia-inducible factor-1α |
| lncRNA | long non-coding RNAs |
| mRNAs | messenger RNAs |
| NPC | nasopharyngeal carcinoma |
| NKILA | NF-B interacting lncRNA |
| ncRNA | non-coding RNA |
| NSCLC | non-small-cell lung carcinoma |
| NF-κB | nuclear factor-B |
| Oct4 | Octamer-binding transcription factor 4 |
| Sox2 | Sex-determining region Y-box2 |
| PDAC | pancreatic ductal adenocarcinoma |
| PI3K | phosphatidylinositol 3-kinase |
| ERK | extracellular signal-regulated kinase |
| MAPK | mitogen-activated protein kinase |
| PTMS | post-translational modifications |
| PRISMA | Preferred Reporting Items for Systemic Reviews and Meta-Analysis |
| PCGEM1 | Prostate cancer gene expression marker |
| RCC | renal cell carcinoma |
| Rnase P | ribonuclease P |
| rRNAs | ribosomal RNAs |
| tRNAs | transfer RNAs |
| STAT3 | signal transducer and activator of transcription 3 |
| EGFR | epidermal growth factor receptor |
| SIRT6 | Sirtuin6 |
| PRC2 | polycomb repressive complex 2 |
| snoRNA | small nucleolar RNA |
| snoRNP | small nucleolar ribonucleoprotein |
| SWI/SNF | Switching defective/sucrose nonfermenting |
References
- Croce, C.M. Oncogenes and Cancer. N. Engl. J. Med. 2008, 358, 502–511. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Tian, S.; Pan, Y.; Li, W.; Wang, Q.; Tang, Y.; Yu, T.; Wu, X.; Shi, Y.; Ma, P.; et al. Pyroptosis: A new frontier in cancer. Biomed. Pharmacother. 2020, 121, 109595. [Google Scholar] [CrossRef]
- Luo, F.; Wen, Y.; Zhou, H.; Li, Z. Roles of long non-coding RNAs in cervical cancer. Life Sci. 2020, 256, 117981. [Google Scholar] [CrossRef]
- Whiteman, D.C.; Wilson, L.F. The fractions of cancer attributable to modifiable factors: A global review. Cancer Epidemiol. 2016, 44, 203–221. [Google Scholar] [CrossRef]
- Luke, J.J.; Flaherty, K.T.; Ribas, A.; Long, G.V. Targeted agents and immunotherapies: Optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol. 2017, 14, 463–482. [Google Scholar] [CrossRef] [Green Version]
- Kensler, T.W.; Spira, A.; Garber, J.E.; Szabo, E.; Lee, J.J.; Dong, Z.; Dannenberg, A.J.; Hait, W.N.; Blackburn, E.; Davidson, N.E.; et al. Transforming Cancer Prevention through Precision Medicine and Immune-oncology. Cancer Prev. Res. 2016, 9, 2–10. [Google Scholar] [CrossRef] [Green Version]
- Collins, F.S.; Varmus, H. A New Initiative on Precision Medicine. N. Engl. J. Med. 2015, 372, 793–795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, A.A.; Letai, A.; Fisher, D.E.; Flaherty, K.T. Precision medicine for cancer with next-generation functional diagnostics. Nat. Rev. Cancer 2015, 15, 747–756. [Google Scholar] [CrossRef]
- Wilusz, J.E.; Sunwoo, H.; Spector, D.L. Long noncoding RNAs: Functional surprises from the RNA world. Genes Dev. 2009, 23, 1494–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Fu, H.; Wu, Y.; Zheng, X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci. China Life Sci. 2013, 56, 876–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Deng, K.; Wang, H.; Xia, J.; Shan, T.; Liang, Z.; Yao, L.; Jin, S. GAS5 Inhibits Gastric Cancer Cell Proliferation Partly by Modulating CDK6. Oncol. Res. Treat. 2015, 38, 362–366. [Google Scholar] [CrossRef]
- Li, S.; Zhou, J.; Wang, Z.; Wang, P.; Gao, X.; Wang, Y. Long noncoding RNA GAS5 suppresses triple negative breast cancer progression through inhibition of proliferation and invasion by competitively binding miR-196a-5p. Biomed. Pharmacother. 2018, 104, 451–457. [Google Scholar] [CrossRef]
- Lee, J.-H.; Parthiban, P.; Jin, G.-Z.; Knowles, J.C.; Kim, H.-W. Materials roles for promoting angiogenesis in tissue regeneration. Prog. Mater. Sci. 2020, 100732. [Google Scholar] [CrossRef]
- Lin, C.-W.; Lin, P.-Y.; Yang, P.-C. Noncoding RNAs in Tumor Epithelial-to-Mesenchymal Transition. Stem Cells Int. 2016, 2016, 2732705. [Google Scholar] [CrossRef] [Green Version]
- Ghafouri-Fard, S.; Shoorei, H.; Mohaqiq, M.; Taheri, M. Non-coding RNAs regulate angiogenic processes. Vasc. Pharmacol. 2020, 133–134, 106778. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, X.; Chen, W.; Hu, X.; Li, J.; Liu, C. Regulatory roles of long noncoding RNAs implicated in cancer hallmarks. Int. J. Cancer 2020, 146, 906–916. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Zhang, Y.; Zhong, Q.; Chen, Q.; Zhang, L. Molecular mechanism of HEIH and HULC in the proliferation and invasion of hepatoma cells. Int. J. Clin. Exp. Med. 2015, 8, 12956–12962. [Google Scholar]
- Wang, F.; Ying, H.-Q.; He, B.-S.; Pan, Y.-Q.; Deng, Q.-W.; Sun, H.-L.; Chen, J.; Liu, X.; Wang, S.-K. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget 2015, 6, 7899–7917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braicu, C.; Mehterov, N.; Vladimirov, B.; Sarafian, V.; Nabavi, S.M.; Atanasov, A.G.; Berindan-Neagoe, I. Nutrigenomics in cancer: Revisiting the effects of natural compounds. Semin. Cancer Biol. 2017, 46, 84–106. [Google Scholar] [CrossRef]
- Król, M.; Pawłowski, K.M.; Majchrzak, K.; Szyszko, K.; Motyl, T. Why chemotherapy can fail? Pol. J. Vet. Sci. 2010, 13, 399–406. [Google Scholar] [PubMed]
- Efferth, T.; Saeed, M.E.; Mirghani, E.; Alim, A.; Yassin, Z.; Saeed, E.; Khalid, H.E.; Daak, S. Integration of phytochemicals and phytotherapy into cancer precision medicine. Oncotarget 2017, 8, 50284–50304. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Yang, Y.; Li, Y.; Cao, Y.; Tang, L.; Chen, F.; Xia, J. Baicalein inhibits cervical cancer progression via downregulating long noncoding RNA BDLNR and its downstream PI3 K/Akt pathway. Int. J. Biochem. Cell Biol. 2018, 94, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Xiang, T.; Wu, Q.-F.; Wang, W.-X. Curcumin suppresses the proliferation of gastric cancer cells by downregulating H19. Oncol. Lett. 2016, 12, 5156–5162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.; Jiang, P.; Zeb, F.; Wu, X.; Xu, C.; Chen, L.; Feng, Q. EGCG regulates CTR1 expression through its pro-oxidative property in non-small-cell lung cancer cells. J. Cell. Physiol. 2020, 235. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Mu, L.; Cui, Y.; Li, Y.; Chen, P.; Xie, H.; Wang, X. Long non-coding RNA CASC2 enhances berberine-induced cytotoxicity in colorectal cancer cells by silencing BCL2. Mol. Med. Rep. 2019, 20, 995–1006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, F.; Li, J.; Ma, C.; Tang, X.; Tang, Q.; Wu, J.; Chai, X.; Xie, J.; Yang, X.; Hann, S.S. Novel regulation of miR-34a-5p and HOTAIR by the combination of berberine and gefitinib leading to inhibition of EMT in human lung cancer. J. Cell. Mol. Med. 2020, 24, 5578–5592. [Google Scholar] [CrossRef]
- Mishra, S.; Verma, S.S.; Rai, V.; Awasthee, N.; Chava, S.; Hui, K.M.; Kumar, A.P.; Challagundla, K.B.; Sethi, G.; Gupta, S.C. Long non-coding RNAs are emerging targets of phytochemicals for cancer and other chronic diseases. Cell. Mol. Life Sci. 2019, 76, 1947–1966. [Google Scholar] [CrossRef]
- Rathinasamy, B.; Velmurugan, B.K. Role of lncRNAs in the cancer development and progression and their regulation by various phytochemicals. Biomed. Pharmacother. 2018, 102, 242–248. [Google Scholar] [CrossRef]
- Saghafi, T.; Taheri, R.A.; Parkkila, S.; Emameh, R.Z. Phytochemicals as Modulators of Long Non-Coding RNAs and Inhibitors of Cancer-Related Carbonic Anhydrases. Int. J. Mol. Sci. 2019, 20, 2939. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Chen, H.; Gao, J.; Liu, Y.; Li, J.; Wang, J. Bioactive Ingredients in Chinese Herbal Medicines That Target Non-coding RNAs: Promising New Choices for Disease Treatment. Front. Pharmacol. 2019, 10, 515. [Google Scholar] [CrossRef] [Green Version]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.D.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA Statement for Reporting Systematic Reviews and Meta-Analyses of Studies That Evaluate Health Care Interventions: Explanation and Elaboration. PLoS Med. 2009, 6, e1000100. [Google Scholar] [CrossRef] [PubMed]
- Dahariya, S.; Paddibhatla, I.; Kumar, S.; Raghuwanshi, S.; Pallepati, A.; Gutti, R.K. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Mol. Immunol. 2019, 112, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, S.U.; Grote, P.; Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.J.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef] [Green Version]
- Vicens, Q.; Westhof, E. Biogenesis of Circular RNAs. Cell 2014, 159, 13–14. [Google Scholar] [CrossRef] [Green Version]
- Hu, G.; Niu, F.; Humburg, B.A.; Liao, K.; Bendi, V.S.; Callen, S.; Fox, H.S.; Buch, S. Molecular mechanisms of long noncoding RNAs and their role in disease pathogenesis. Oncotarget 2018, 9, 18648–18663. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Wang, J.; Lian, Y.; Fan, C.; Zhang, P.; Wu, Y.; Li, X.; Xiong, F.; Li, X.; Li, G.; et al. Linking long non-coding RNAs and SWI/SNF complexes to chromatin remodeling in cancer. Mol. Cancer 2017, 16, 42. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.S.; Roberts, C.W.M. Linking the SWI/SNF complex to prostate cancer. Nat. Genet. 2013, 45, 1268–1269. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Lian, J.B.; Stein, J.L.; Stein, G.S.; Nickerson, A.J.; Imbalzano, A.N. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics 2017, 9, 919–931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Mueller, M.; Geng, T.; Shen, Y.; Liu, Y.; Hou, P.; Ramillapalli, R.; Taylor, H.S.; Paidas, M.; Huang, Y. H19 lncRNA alters methylation and expression of Hnf4α in the liver of metformin-exposed fetuses. Cell Death Dis. 2017, 8, e3175. [Google Scholar] [CrossRef]
- Jain, A.K.; Xi, Y.; McCarthy, R.; Allton, K.; Akdemir, K.C.; Patel, L.R.; Aronow, B.; Lin, C.; Li, W.; Yang, L.; et al. LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol. Cell 2016, 64, 967–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, M.; Chen, X.; Lin, K.; Zeng, K.; Liu, X.; Pan, B.; Xu, X.; Xu, T.; Hu, X.; Sun, L.; et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol. Cancer 2018, 17, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.M.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nat. Cell Biol. 2009, 458, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.Y.; Do, B.T.; Webster, E.D.; Khavari, P.A.; Chang, H.Y. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat. Struct. Mol. Biol. 2014, 21, 585–590. [Google Scholar] [CrossRef] [Green Version]
- Peng, W.-X.; Koirala, P.; Mo, Y.-Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef]
- Guan, H.; Zhu, T.; Wu, S.; Liu, S.; Liu, B.; Wu, J.; Cai, J.; Zhu, X.; Zhang, X.; Zeng, M.; et al. Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/β-catenin signaling. Proc. Natl. Acad. Sci. USA 2019, 116, 14019–14028. [Google Scholar] [CrossRef] [Green Version]
- Kane, A.B.; Jean, D.; Knuutila, S.; Jaurand, M.-C. Malignant Mesothelioma: Mechanism of Carcinogenesis. In Occupational Cancers; Springer: Cham, Switzerland, 2020; pp. 343–362. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, E.-B.; Kong, R.; Yin, D.-D.; You, L.-H.; Sun, M.; Han, L.; Xu, T.-P.; Xia, R.; Yang, J.-S.; De, W.; et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget 2014, 5, 2276–2292. [Google Scholar] [CrossRef] [Green Version]
- Nie, F.-Q.; Sun, M.; Yang, J.-S.; Xie, M.; Xu, T.-P.; Xia, R.; Liu, Y.-W.; Liu, X.-H.; Zhang, E.-B.; Lu, K.-H.; et al. Long Noncoding RNA ANRIL Promotes Non–Small Cell Lung Cancer Cell Proliferation and Inhibits Apoptosis by Silencing KLF2 and P21 Expression. Mol. Cancer Ther. 2015, 14, 268–277. [Google Scholar] [CrossRef] [Green Version]
- Naemura, M.; Murasaki, C.; Inoue, Y.; Okamoto, H.; Kotake, Y. Long Noncoding RNA ANRIL Regulates Proliferation of Non-small Cell Lung Cancer and Cervical Cancer Cells. Anticancer Res. 2015, 35, 5377–5382. [Google Scholar]
- Ma, J.; Li, T.; Han, X.; Yuan, H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2018, 144, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Hua, Y. CCAT1: An oncogenic long noncoding RNA in human cancers. J. Cancer Res. Clin. Oncol. 2016, 143, 555–562. [Google Scholar] [CrossRef]
- Redis, R.S.; Vela, L.E.; Lu, W.; De Oliveira, J.F.; Ivan, C.; Rodriguez-Aguayo, C.; Adamoski, D.; Pasculli, B.; Taguchi, A.; Chen, Y.; et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol. Cell 2016, 61, 640. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.; Zambalde, E.; Mathias, C.; Barazetti, J.; Gradia, D.; Oliveira, J. lncRNAs in Hallmarks of Cancer and Clinical Applications. In Non-Coding RNAs; InTechOpen: London, UK, 2019. [Google Scholar]
- Ellis, B.C.; Graham, L.D.; Molloy, P.L. CRNDE, a long non-coding RNA responsive to insulin/IGF signaling, regulates genes involved in central metabolism. Biochim. Biophys. Acta 2014, 1843, 372–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Tang, Y.; Xing, W.; Dong, W.; Wang, Z. LncRNA, CRNDE promotes osteosarcoma cell proliferation, invasion and migration by regulating Notch1 signaling and epithelial-mesenchymal transition. Exp. Mol. Pathol. 2018, 104, 19–25. [Google Scholar] [CrossRef]
- Chang, L.; Li, C.; Lan, T.; Wu, L.; Yuan, Y.; Liu, Q.; Liu, Z. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol. Med. Rep. 2016, 13, 1541–1550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, W.P.; Ng, E.K.; Ng, S.S.; Jin, H.; Yu, J.; Sung, J.J.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2009, 31, 350–358. [Google Scholar] [CrossRef]
- Liang, W.-C.; Fu, W.-M.; Wong, C.-W.; Wang, Y.; Wang, W.-M.; Hu, G.-X.; Zhang, L.; Xiao, L.-J.; Wan, D.C.-C.; Zhang, J.-F.; et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget 2015, 6, 22513–22525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, J.; Mo, J.; Luo, M.; Yu, Q.; Zhou, S.; Li, T.; Zhang, Y.; Luo, W. c-Myc-activated long non-coding RNA H19 downregulates miR-107 and promotes cell cycle progression of non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 12400–12409. [Google Scholar] [PubMed]
- Fu, W.-M.; Lu, Y.-F.; Hu, B.-G.; Liang, W.-C.; Zhu, X.; Yang, H.-D.; Li, G.; Zhang, J.-F. Long noncoding RNA hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2016, 7, 4712–4723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, J.-J.; Wang, Y.; Ding, J.-X.; Jing-Xin, D.; Yang, G.; Hua, K.-Q. The long non-coding RNA HOTAIR promotes the proliferation of serous ovarian cancer cells through the regulation of cell cycle arrest and apoptosis. Exp. Cell Res. 2015, 333, 238–248. [Google Scholar] [CrossRef]
- Feng, T.; Shao, F.; Wu, Q.; Zhang, X.; Xu, D.; Qian, K.; Xie, Y.; Wang, S.; Xu, N.; Wang, Y.; et al. miR-124 downregulation leads to breast cancer progression via LncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget 2016, 7, 16205–16216. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; He, H.; Xiao, W.; Liu, Q.; Deng, Z.; Lu, Y.; Wang, Q.; Zheng, Q.; Li, Y. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget 2016, 7, 54733–54743. [Google Scholar] [CrossRef] [Green Version]
- Shi, B.; Wang, Y.; Yin, F. MALAT1/miR-124/Capn4 axis regulates proliferation, invasion and EMT in nasopharyngeal carcinoma cells. Cancer Biol. Ther. 2017, 18, 792–800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Liang, L.; Ouyang, K.; Li, Z.; Yi, X. MALAT1 induces tongue cancer cells’ EMT and inhibits apoptosis through Wnt/β-catenin signaling pathway. J. Oral Pathol. Med. 2017, 46, 98–105. [Google Scholar] [CrossRef]
- Liu, J.; Peng, W.-X.; Mo, Y.-Y.; Luo, D. MALAT1-mediated tumorigenesis. Front. Biosci. (Landmark Ed.) 2017, 22, 66–80. [Google Scholar] [PubMed]
- Guo, Q.; Qian, Z.; Yan, D.; Li, L.; Huang, L. LncRNA-MEG3 inhibits cell proliferation of endometrial carcinoma by repressing Notch signaling. Biomed. Pharmacother. 2016, 82, 589–594. [Google Scholar] [CrossRef]
- Lu, K.-H.; Li, W.; Liu, X.-H.; Sun, M.; Zhang, M.-L.; Wu, W.-Q.; Xie, W.-P.; Hou, Y.-Y. Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 2013, 13, 461. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Chi, G.; Zhao, C.; Li, D. Long noncoding RNA MEG3 inhibits proliferation and migration but induces autophagy by regulation of Sirt7 and PI3K/AKT/mTOR pathway in glioma cells. J. Cell. Biochem. 2019, 120, 7516–7526. [Google Scholar] [CrossRef]
- Tseng, Y.-Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nat. Cell Biol. 2014, 512, 82–86. [Google Scholar] [CrossRef]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Bettin, N.; Pegorar, C.O.; Cusanelli, E. The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability. Cells 2019, 8, 246. [Google Scholar] [CrossRef] [Green Version]
- Redon, S.; Reichenbach, P.; Lingner, J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010, 38, 5797–5806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, M.; Pang, H.; Li, X.; Li, H.; Pan, J.; Chen, W. Long non-coding RNA urothelial cancer-associated 1 promotes bladder cancer cell migration and invasion by way of the hsa-miR-145– ZEB 1/2– FSCN 1 pathway. Cancer Sci. 2015, 107, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Cai, W.; Ren, S.; Li, X.; Wang, Q.; Pan, H.; Zhao, M.; Li, J.; Zhang, Y.; Zhao, C.; et al. Long non-coding RNA UCA1 induces non-T790M acquired resistance to EGFR-TKIs by activating the AKT/mTOR pathway in EGFR-mutant non-small cell lung cancer. Oncotarget 2015, 6, 23582–23593. [Google Scholar] [CrossRef] [Green Version]
- Dhamija, S.; Diederichs, S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int. J. Cancer 2016, 139, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.-D.; Liu, X.-Y.; Lin, Y.; Liu, H.-F.; Zhang, G.-J. LncRNA CASC9 promotes tumorigenesis by affecting EMT and predicts poor prognosis in esophageal squamous cell cancer. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 422–429. [Google Scholar]
- Khan, H.; Ullah, H.; Castilho, P.C.M.F.; Gomila, A.S.; D’Onofrio, G.; Filosa, R.; Wang, F.; Nabavi, S.M.; Daglia, M.; Silva, A.S.; et al. Targeting NF-κB signaling pathway in cancer by dietary polyphenols. Crit. Rev. Food Sci. Nutr. 2019, 60, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Li, J.; Wang, P.; Zhang, Z.; Wang, X. LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF-κB. J. Cell. Biochem. 2019, 120, 19415–19421. [Google Scholar] [CrossRef]
- Yang, T.; Li, S.; Liu, J.; Yin, D.; Yang, X.; Tang, Q. lncRNA-NKILA/NF-κB feedback loop modulates laryngeal cancer cell proliferation, invasion, and radioresistance. Cancer Med. 2018, 7, 2048–2063. [Google Scholar] [CrossRef]
- Li, P.; Zhang, X.; Wang, L.; Du, L.; Yang, Y.; Liu, T.; Li, C.; Wang, C. lncRNA HOTAIR Contributes to 5FU Resistance through Suppressing miR-218 and Activating NF-κB/TS Signaling in Colorectal Cancer. Mol. Ther. Nucleic Acids 2020, 20, 879–880. [Google Scholar] [CrossRef]
- Salamati, A.; Majidinia, M.; Asemi, Z.; Sadeghpour, A.; Oskoii, M.A.; Shanebandi, D.; Alemi, F.; Mohammadi, E.; Karimian, A.; Targhazeh, N.; et al. Modulation of telomerase expression and function by miRNAs: Anti-cancer potential. Life Sci. 2020, 259, 118387. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.-F.; Tang, L.; Ouyang, W.-X.; Jiang, T.; Zhang, H.; Li, S.-J. β-catenin-coordinated lncRNA MALAT1 up-regulation of ZEB-1 could enhance the telomerase activity in HGF-mediated differentiation of bone marrow mesenchymal stem cells into hepatocytes. Pathol. Res. Pract. 2019, 215, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Z.; Duan, Y. LncRNA MEG3 inhibits non-small cell lung cancer via interaction with DKC1 protein. Oncol. Lett. 2020, 20, 2183–2190. [Google Scholar] [CrossRef]
- Kroustallaki, P.; Lirussi, L.; Carracedo, S.; You, P.; Esbensen, Q.Y.; Götz, A.; Jobert, L.; Alsøe, L.; Sætrom, P.; Gagos, S.; et al. SMUG1 Promotes Telomere Maintenance through Telomerase RNA Processing. Cell Rep. 2019, 28, 1690–1702.e10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.-H. Crosstalk of lncRNA and Cellular Metabolism and Their Regulatory Mechanism in Cancer. Int. J. Mol. Sci. 2020, 21, 2947. [Google Scholar] [CrossRef]
- Munn, Z.; Tufanaru, C.; Aromataris, E. Recognition of the health assistant as a delegated clinical role and their inclusion in models of care: A systematic review and meta-synthesis of qualitative evidence. Int. J. Evid. Based Healthc. 2013, 11, 3–19. [Google Scholar] [CrossRef]
- Xue, M.; Li, X.; Li, Z.; Chen, W. Urothelial carcinoma associated 1 is a hypoxia-inducible factor-1α-targeted long noncoding RNA that enhances hypoxic bladder cancer cell proliferation, migration, and invasion. Tumor Biol. 2014, 35, 6901–6912. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Wang, Z.; Hu, Q.; Wu, J.; Li, Y.; Ren, X.; Wu, T.; Tao, X.; Chen, X.; et al. LncRNA-p23154 promotes the invasion-metastasis potential of oral squamous cell carcinoma by regulating Glut1-mediated glycolysis. Cancer Lett. 2018, 434, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, Y.; Lin, L.; Huang, Q.; He, W.; Zhang, S.; Dong, S.; Wen, Z.; Rao, J.; Liao, W.; et al. The lncRNA MACC1-AS1 promotes gastric cancer cell metabolic plasticity via AMPK/Lin28 mediated mRNA stability of MACC1. Mol. Cancer 2018, 17, 69. [Google Scholar] [CrossRef]
- Xiao, Q.; Zhou, Y.; Winter, S.; Büttner, F.; Schaeffeler, E.; Schwab, M.; Lauschke, V.M. Germline variant burden in multidrug resistance transporters is a therapy-specific predictor of survival in breast cancer patients. Int. J. Cancer 2020, 146, 2475–2487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.K.; Singh, P.; Ali, V.; Verma, M. Role of membrane-embedded drug efflux ABC transporters in the cancer chemotherapy. Oncol. Rev. 2020, 14, 448. [Google Scholar] [CrossRef]
- Huang, P.; Ouyang, D.-J.; Chang, S.; Li, M.-Y.; Li, L.; Li, Q.-Y.; Zeng, R.; Zou, Q.-Y.; Su, J.; Zhao, P.; et al. Chemotherapy-driven increases in the CDKN1A/PTN/PTPRZ1 axis promote chemoresistance by activating the NF-κB pathway in breast cancer cells. Cell Commun. Signal. 2018, 16, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.-T.; Han, C.; Sun, Y.-M.; Chen, T.-Q.; Chen, Y.-Q. Noncoding RNAs in cancer therapy resistance and targeted drug development. J. Hematol. Oncol. 2019, 12, 55. [Google Scholar] [CrossRef]
- Liu, J.; Wan, L.; Lu, K.; Sun, M.; Pan, X.; Zhang, P.; Lu, B.; Liu, G.; Wang, Z. The Long Noncoding RNA MEG3 Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma. PLoS ONE 2015, 10, e0114586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Zhu, Y. Effect of lncRNA ANRIL knockdown on proliferation and cisplatin chemoresistance of osteosarcoma cells in vitro. Pathol. Res. Pract. 2019, 215, 931–938. [Google Scholar] [CrossRef]
- Xu, R.; Mao, Y.; Chen, K.; He, W.; Shi, W.; Han, Y. The long noncoding RNA ANRIL acts as an oncogene and contributes to paclitaxel resistance of lung adenocarcinoma A549 cells. Oncotarget 2017, 8, 39177–39184. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Sun, M.; Lu, K.; Liu, J.; Zhang, M.; Wu, W.; De, W.; Wang, Z.; Wang, R. The Long Noncoding RNA HOTAIR Contributes to Cisplatin Resistance of Human Lung Adenocarcinoma Cells via downregualtion of p21WAF1/CIP1 Expression. PLoS ONE 2013, 8, e77293. [Google Scholar] [CrossRef]
- Liu, M.-Y.; Li, X.-Q.; Gao, T.-H.; Cui, Y.; Ma, N.; Zhou, Y.; Zhang, G.-J. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J. Thorac. Dis. 2016, 8, 3314–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Cao, Z.; Guo, H.; Li, S. The action mechanism of lncRNA-HOTAIR on the drug resistance of non-small cell lung cancer by regulating Wnt signaling pathway. Exp. Ther. Med. 2018, 15, 4885–4889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Xia, J.; Xie, S.; Zou, R.; Pan, S.; Wang, Z.-W.; Assaraf, Y.G.; Zhu, X. Long non-coding RNAs as a determinant of cancer drug resistance: Towards the overcoming of chemoresistance via modulation of lncRNAs. Drug Resist. Updat. 2020, 50, 100683. [Google Scholar] [CrossRef]
- Wang, H.-L.V.; Chekanova, J.A. Long Noncoding RNAs in Plants. Adv. Exp. Med. Biol. 2017, 1008, 133–154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Ji, B.; Liu, K.; Hu, G.; Wang, F.; Chen, Q.; Yu, R.; Huang, P.; Ren, J.; Guo, C.; et al. EVLncRNAs 2.0: An updated database of manually curated functional long non-coding RNAs validated by low-throughput experiments. Nucleic Acids Res. 2021, 49, D86–D91. [Google Scholar] [CrossRef] [PubMed]
- Crespi, M.D.; Jurkevitch, E.; Poiret, M.; d’Aubenton-Carafa, Y.; Petrovics, G.; Kondorosi, E.; Kondorosi, A. enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J. 1994, 13, 5099–5112. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, Q.-H.; Kaufmann, K. Long non-coding RNAs in plants: Emerging modulators of gene activity in development and stress responses. Planta 2020, 252, 92. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, J.; Deng, F.; Wang, W.; Cheng, Y.; Song, L.; Hu, M.; Shen, J.; Xu, Q.; Shen, F. The long non-coding RNA lncRNA973 is involved in cotton response to salt stress. BMC Plant Biol. 2019, 19, 459. [Google Scholar] [CrossRef] [PubMed]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [Green Version]
- Kashyap, D.; Tuli, H.S.; Yerer, M.B.; Sharma, A.; Sak, K.; Srivastava, S.; Pandey, A.; Garg, V.K.; Sethi, G.; Bishayee, A. Natural product-based nanoformulations for cancer therapy: Opportunities and challenges. Semin. Cancer Biol. 2021, 69, 5–23. [Google Scholar] [CrossRef]
- Dillard, C.J.; German, J.B. Phytochemicals: Nutraceuticals and human health. J. Sci. Food Agric. 2000, 80, 1744–1756. [Google Scholar] [CrossRef]
- Tewari, D.; Patni, P.; Bishayee, A.; Sah, A.N.; Bishayee, A. Natural products targeting the PI3K-Akt-mTOR signaling pathway in cancer: A novel therapeutic strategy. Semin. Cancer Biol. 2019. [Google Scholar] [CrossRef]
- Schultz, D.J.; Krishna, A.; Vittitow, S.L.; Alizadeh-Rad, N.; Muluhngwi, P.; Rouchka, E.C.; Klinge, C.M. Transcriptomic response of breast cancer cells to anacardic acid. Sci. Rep. 2018, 8, 8063. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Jiang, J.; Zhang, C.; Li, Y. Baicalein restrains proliferation, migration, and invasion of human malignant melanoma cells by down-regulating colon cancer associated transcript-1. Braz. J. Med. Biol. Res. 2019, 52, 8934. [Google Scholar] [CrossRef]
- Yu, X.; Tang, W.; Yang, Y.; Tang, L.; Dai, R.; Pu, B.; Feng, C.; Xia, J. Long noncoding RNA NKILA enhances the anti-cancer effects of baicalein in hepatocellular carcinoma via the regulation of NF-κB signaling. Chem. Interact. 2018, 285, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Cao, Y.; Tang, L.; Yang, Y.; Chen, F.; Xia, J. Baicalein inhibits breast cancer growth via activating a novel isoform of the long noncoding RNA PAX8-AS1-N. J. Cell. Biochem. 2018, 119, 6842–6856. [Google Scholar] [CrossRef]
- Dai, W.; Mu, L.; Cui, Y.; Li, Y.; Chen, P.; Xie, H.; Wang, X. Berberine Promotes Apoptosis of Colorectal Cancer via Regulation of the Long Non-Coding RNA (lncRNA) Cancer Susceptibility Candidate 2 (CASC2)/AU-Binding Factor 1 (AUF1)/B-Cell CLL/Lymphoma 2 (Bcl-2) Axis. Med. Sci. Monit. 2019, 25, 730–738. [Google Scholar] [CrossRef] [PubMed]
- Awasthee, N.; Rai, V.; Verma, S.S.; Francis, K.S.; Nair, M.S.; Gupta, S.C. Anti-cancer activities of Bharangin against breast cancer: Evidence for the role of NF-κB and lncRNAs. Biochim. Biophys. Acta 2018, 1862, 2738–2749. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, Y.; Zhang, X.; Ren, Q.; Huiling, L.; Huang, Y.; Lu, H.; Chen, J. Calycosin inhibits the in vitro and in vivo growth of breast cancer cells through WDR7-7-GPR30 Signaling. J. Exp. Clin. Cancer Res. 2017, 36, 153. [Google Scholar] [CrossRef]
- Chen, J.; Lin, C.; Yong, W.; Ye, Y.; Huang, Z. Calycosin and Genistein Induce Apoptosis by Inactivation of HOTAIR/p-Akt Signaling Pathway in Human Breast Cancer MCF-7 Cells. Cell. Physiol. Biochem. 2015, 35, 722–728. [Google Scholar] [CrossRef]
- Kong, L.; Li, X.; Wang, H.; He, G.; Tang, A. Calycosin inhibits nasopharyngeal carcinoma cells by influencing EWSAT1 expression to regulate the TRAF6-related pathways. Biomed. Pharmacother. 2018, 106, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Toden, S.; Ravindranathan, P.; Han, H.; Goel, A. Curcumin sensitizes pancreatic cancer cells to gemcitabine by attenuating PRC2 subunit EZH2, and the lncRNA PVT1 expression. Carcinogenesis 2017, 38, 1036–1046. [Google Scholar] [CrossRef] [Green Version]
- Garitano-Trojaola, A.; José-Eneriz, E.S.; Ezponda, T.; Unfried, J.P.; Carrasco-León, A.; Razquin, N.; Barriocanal, M.; Vilas-Zornoza, A.; Sangro, B.; Segura, V.; et al. Deregulation of linc-PINT in acute lymphoblastic leukemia is implicated in abnormal proliferation of leukemic cells. Oncotarget 2018, 9, 12842–12852. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Chi, H.; Chen, J.; Chen, C.; Huang, Y.; Xi, H.; Xue, J.; Si, Y. Curcumin suppresses proliferation and in vitro invasion of human prostate cancer stem cells by ceRNA effect of miR-145 and lncRNA-ROR. Gene 2017, 631, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Pei, C.-S.; Wu, H.-Y.; Fan, F.-T.; Wu, Y.; Shen, C.-S.; Pan, L.-Q. Influence of Curcumin on HOTAIR-Mediated Migration of Human Renal Cell Carcinoma Cells. Asian Pac. J. Cancer Prev. 2014, 15, 4239–4243. [Google Scholar] [CrossRef] [Green Version]
- Sun, K.; Jia, Z.; Duan, R.; Yan, Z.; Jin, Z.; Yan, L.; Li, Q.; Yang, J. Long non-coding RNA XIST regulates miR-106b-5p/P21 axis to suppress tumor progression in renal cell carcinoma. Biochem. Biophys. Res. Commun. 2019, 510, 416–420. [Google Scholar] [CrossRef]
- Esmatabadi, M.J.D.; Motamedrad, M.; Sadeghizadeh, M. Down-regulation of lncRNA, GAS5 decreases chemotherapeutic effect of dendrosomal curcumin (DNC) in breast cancer cells. Phytomedicine 2018, 42, 56–65. [Google Scholar] [CrossRef]
- Yu, H.; Xie, Y.; Zhou, Z.; Wu, Z.; Dai, X.; Xu, B. Curcumin Regulates the Progression of Colorectal Cancer via LncRNA NBR2/AMPK Pathway. Technol. Cancer Res. Treat. 2019, 18, 1533033819870781. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Sadeghizadeh, M.; Behmanesh, M.; Najafi, F. Dendrosomal curcumin increases expression of the long non-coding RNA gene MEG3 via up-regulation of epi-miRs in hepatocellular cancer. Phytomedicine 2015, 22, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, J.; Xu, X.; Li, L. Curcumin suppresses cisplatin resistance development partly via modulating extracellular vesicle-mediated transfer of MEG3 and miR-214 in ovarian cancer. Cancer Chemother. Pharmacol. 2017, 79, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-H.; Chen, J.; Zhang, B.-R.; Lu, S.-J.; Wang, F.; Peng, L.; Dai, J.-H.; Sun, Y.-Z. Curcumin inhibits proliferation and enhances apoptosis in A549 cells by downregulating lncRNA UCA1. Die Pharm. 2018, 73, 402–407. [Google Scholar]
- Ho, T.-T.; Huang, J.; Zhou, N.; Zhang, Z.; Koirala, P.; Zhou, X.; Wu, F.; Ding, X.; Mo, Y.-Y. Regulation of PCGEM1 by p54/nrb in prostate cancer. Sci. Rep. 2016, 6, 34529. [Google Scholar] [CrossRef] [Green Version]
- Zinovieva, O.L.; Grineva, E.N.; Prokofjeva, M.M.; Karpov, D.S.; Krasnov, G.S.; Prassolov, V.S.; Mashkova, T.D.; Lisitsyn, N.A. Treatment with anti-cancer agents results in profound changes in lncRNA expression in colon cancer cells. Mol. Biol. 2017, 51, 841–848. [Google Scholar] [CrossRef]
- Jiang, P.; Wu, X.; Wang, X.; Huang, W.; Feng, Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget 2016, 7, 43337–43351. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Chen, D.; Zhu, K. SOX2OT variant 7 contributes to the synergistic interaction between EGCG and Doxorubicin to kill osteosarcoma via autophagy and stemness inhibition. J. Exp. Clin. Cancer Res. 2018, 37, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Chen, X.; Jiang, J.; Wan, X.; Wang, Y.; Xu, P. Epigallocatechin gallate reverses gastric cancer by regulating the long noncoding RNA LINC00511/miR-29b/KDM2A axis. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165856. [Google Scholar] [CrossRef]
- Liu, D.; Li, Y.; Luo, G.; Xiao, X.; Tao, D.; Wu, X.; Wang, M.; Huang, C.; Wang, L.; Zeng, F.; et al. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017, 388, 281–291. [Google Scholar] [CrossRef]
- Wang, M.; Guo, C.; Wang, L.; Luo, G.; Huang, C.; Li, Y.; Liu, D.; Zeng, F.; Jiang, G.; Xiao, X. Long noncoding RNA GAS5 promotes bladder cancer cells apoptosis through inhibiting EZH2 transcription. Cell Death Dis. 2018, 9, 238. [Google Scholar] [CrossRef] [PubMed]
- Chiyomaru, T.; Fukuhara, S.; Saini, S.; Majid, S.; Deng, G.; Shahryari, V.; Chang, I.; Tanaka, Y.; Enokida, H.; Nakagawa, M.; et al. Long Non-coding RNA HOTAIR Is Targeted and Regulated by miR-141 in Human Cancer Cells. J. Biol. Chem. 2014, 289, 12550–12565. [Google Scholar] [CrossRef] [Green Version]
- Chiyomaru, T.; Yamamura, S.; Fukuhara, S.; Yoshino, H.; Kinoshita, T.; Majid, S.; Saini, S.; Chang, I.; Tanaka, Y.; Enokida, H.; et al. Genistein Inhibits Prostate Cancer Cell Growth by Targeting miR-34a and Oncogenic HOTAIR. PLoS ONE 2013, 8, e70372. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, Y.; Gu, J.; Liang, P.; Shen, M.; Xi, J.; Qin, J. Anti-invasive effect and pharmacological mechanism of genistein against colorectal cancer. BioFactors 2020, 46, 620–628. [Google Scholar] [CrossRef]
- Jeong, D.; Ham, J.; Park, S.; Kim, H.W.; Kim, H.; Ji, H.W.; Kim, S.J. Ginsenoside Rh2 Suppresses Breast Cancer Cell Proliferation by Epigenetically Regulating the Long Noncoding RNA C3orf67-AS1. Am. J. Chin. Med. 2019, 47, 1643–1658. [Google Scholar] [CrossRef]
- Ham, J.; Jeong, D.; Park, S.; Kim, H.W.; Kim, H.; Kim, S.J. Ginsenoside Rg3 and Korean Red Ginseng extract epigenetically regulate the tumor-related long noncoding RNAs RFX3-AS1 and STXBP5-AS1. J. Ginseng Res. 2019, 43, 625–634. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Su, H.; Zou, C.; Liang, X.; Fei, Z. Ginsenoside Rg3 suppresses the growth of gemcitabine-resistant pancreatic cancer cells by upregulating lncRNA-CASC2 and activating PTEN signaling. J. Biochem. Mol. Toxicol. 2020, 34, e22480. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Qi, Y. Ginsenoside Rg3 inhibits cell growth, migration and invasion in Caco-2 cells by downregulation of lncRNA CCAT1. Exp. Mol. Pathol. 2019, 106, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhou, Y.; Chen, W.; Chen, L.; Lu, J.; He, F.; Li, X.; Zhao, L. Ginsenoside 20(S)-Rg3 Prevents PKM2-Targeting miR-324-5p from H19 Sponging to Antagonize the Warburg Effect in Ovarian Cancer Cells. Cell. Physiol. Biochem. 2018, 51, 1340–1353. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zhao, P.; Xu, H. Hyperoside exhibits anticancer activity in non-small cell lung cancer cells with T790M mutations by upregulating FoxO1 via CCAT1. Oncol. Rep. 2019, 43, 617–624. [Google Scholar] [CrossRef]
- Liu, C.; Lin, Y.; Xu, J.; Chu, H.; Hao, S.; Liu, X.; Song, X.; Jiang, L.; Zheng, H. Luteolin suppresses tumor progression through lncRNA BANCR and its downstream TSHR/CCND1 signaling in thyroid carcinoma. Int. J. Clin. Exp. Pathol. 2017, 10, 9591–9598. [Google Scholar]
- Hu, T.; Fei, Z.; Su, H.; Xie, R.; Chen, L. Polydatin inhibits proliferation and promotes apoptosis of doxorubicin-resistant osteosarcoma through LncRNA TUG1 mediated suppression of Akt signaling. Toxicol. Appl. Pharmacol. 2019, 371, 55–62. [Google Scholar] [CrossRef]
- Lu, X.; Chen, D.; Yang, F.; Xing, N. Quercetin Inhibits Epithelial-to-Mesenchymal Transition (EMT) Process and Promotes Apoptosis in Prostate Cancer via Downregulating lncRNA MALAT1. Cancer Manag. Res. 2020, 12, 1741–1750. [Google Scholar] [CrossRef] [Green Version]
- Al Aameri, R.F.H.; Sheth, S.; Alanisi, E.M.A.; Borse, V.; Mukherjea, D.; Rybak, L.P.; Ramkumar, V. Tonic suppression of PCAT29 by the IL-6 signaling pathway in prostate cancer: Reversal by resveratrol. PLoS ONE 2017, 12, e0177198. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Guo, X.; Zhang, L.; Ma, Y.; Wang, L.; Liu, Z.; Ji, H.; Xiong, Y. Resveratrol inhibits proliferation, migration and invasion of multiple myeloma cells via NEAT1-mediated Wnt/β-catenin signaling pathway. Biomed. Pharmacother. 2018, 107, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Liu, X.; Fu, X.; Zhang, L.; Sui, H.; Zhou, L.; Sun, J.; Cai, J.; Qin, J.; Ren, J.; et al. Resveratrol Inhibits Invasion and Metastasis of Colorectal Cancer Cells via MALAT1 Mediated Wnt/β-Catenin Signal Pathway. PLoS ONE 2013, 8, e78700. [Google Scholar] [CrossRef] [PubMed]
- Tomita, S.; Abdalla, M.O.A.; Fujiwara, S.; Matsumori, H.; Maehara, K.; Ohkawa, Y.; Iwase, H.; Saitoh, N.; Nakao, M. A cluster of noncoding RNAs activates the ESR1 locus during breast cancer adaptation. Nat. Commun. 2015, 6, 6966. [Google Scholar] [CrossRef]
- Yang, Q.; Xu, E.; Dai, J.; Liu, B.; Han, Z.; Wu, J.; Zhang, S.; Peng, B.; Zhang, Y.; Jiang, Y. A novel long noncoding RNA AK001796 acts as an oncogene and is involved in cell growth inhibition by resveratrol in lung cancer. Toxicol. Appl. Pharmacol. 2015, 285, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Leng, T.; Zhang, Q.; Zhao, Q.; Nie, X.; Yang, L. Sanguinarine inhibits epithelial ovarian cancer development via regulating long non-coding RNA CASC2-EIF4A3 axis and/or inhibiting NF-κB signaling or PI3K/AKT/mTOR pathway. Biomed. Pharmacother. 2018, 102, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Imai-Sumida, M.; Chiyomaru, T.; Majid, S.; Saini, S.; Nip, H.; Dahiya, R.; Tanaka, Y.; Yamamura, S. Silibinin suppresses bladder cancer through down-regulation of actin cytoskeleton and PI3K/Akt signaling pathways. Oncotarget 2017, 8, 92032–92042. [Google Scholar] [CrossRef] [PubMed]
- Beaver, L.M.; Kuintzle, R.; Buchanan, A.; Wiley, M.W.; Glasser, S.T.; Wong, C.P.; Johnson, G.S.; Chang, J.H.; Löhr, C.V.; Williams, D.E.; et al. Long noncoding RNAs and sulforaphane: A target for chemoprevention and suppression of prostate cancer. J. Nutr. Biochem. 2017, 42, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Johnson, G.S.; Li, J.; Beaver, L.M.; Dashwood, W.M.; Sun, D.; Rajendran, P.; Williams, D.E.; Ho, E.; Dashwood, R.H. A functional pseudogene, NMRAL2P, is regulated by Nrf2 and serves as a coactivator ofNQO1in sulforaphane-treated colon cancer cells. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [Green Version]
- Schultz, D.J.; Wickramasinghe, N.S.; Ivanova, M.M.; Isaacs, S.M.; Dougherty, S.M.; Imbert-Fernandez, Y.; Cunningham, A.R.; Chen, C.; Klinge, C.M. Anacardic Acid Inhibits Estrogen Receptor α–DNA Binding and Reduces Target Gene Transcription and Breast Cancer Cell Proliferation. Mol. Cancer Ther. 2010, 9, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Han, E.; Xing, Q.; Yan, J.; Arrington, A.; Wang, C.; Tully, D.; Kowolik, C.M.; Lu, D.M.; Frankel, P.H.; et al. Baicalein upregulates DDIT4 expression which mediates mTOR inhibition and growth inhibition in cancer cells. Cancer Lett. 2015, 358, 170–179. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Yu, Y.; Xu, B.; Zhang, M.; Li, Q.; Miao, L. Pivotal prognostic and diagnostic role of the long non-coding RNA colon cancer-associated transcript 1 expression in human cancer (Review). Mol. Med. Rep. 2018, 19, 771–782. [Google Scholar] [CrossRef] [Green Version]
- Piyanuch, R.; Sukhthankar, M.; Wandee, G.; Baek, S.J. Berberine, a natural isoquinoline alkaloid, induces NAG-1 and ATF3 expression in human colorectal cancer cells. Cancer Lett. 2007, 258, 230–240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, G.; Wu, X.; Li, S.; Xu, X.; Zhu, H.; Chen, X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci. Rep. 2016, 6, 26524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, X.; Alsager, S.; Zhuo, Y.; Shan, B. HOX transcript antisense RNA (HOTAIR) in cancer. Cancer Lett. 2019, 454, 90–97. [Google Scholar] [CrossRef]
- Khatoon, E.; Banik, K.; Harsha, C.; Sailo, B.L.; Thakur, K.K.; Khwairakpam, A.D.; Vikkurthi, R.; Devi, T.B.; Gupta, S.C.; Kunnumakkara, A.B. Phytochemicals in cancer cell chemosensitization: Current knowledge and future perspectives. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Wang, X.; Guo, S.; Xiao, L.; Liang, C.; Wang, Z.; Li, Y.; Liu, Y.; Yao, R.; Liu, Y.; et al. LncRNA HOTAIR functions as a competing endogenous RNA to upregulate SIRT1 by sponging miR-34a in diabetic cardiomyopathy. J. Cell. Physiol. 2019, 234, 4944–4958. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Kannappan, R.; Kim, J.; Rahman, G.M.; Francis, S.K.; Raveendran, R.; Nair, M.S.; Das, J.; Aggarwal, B.B. Bharangin, a Diterpenoid Quinonemethide, Abolishes Constitutive and Inducible Nuclear Factor-κB (NF-κB) Activation by Modifying p65 on Cysteine 38 Residue and Reducing Inhibitor of Nuclear Factor-κB α Kinase Activation, Leading to Suppression of NF-κB-Regulated Gene Expression and Sensitization of Tumor Cells to Chemotherapeutic Agents. Mol. Pharmacol. 2011, 80, 769–781. [Google Scholar] [CrossRef] [Green Version]
- Křížová, L.; Dadáková, K.; Kašparovská, J.; Kašparovský, T. Isoflavones. Molecules 2019, 24, 1076. [Google Scholar] [CrossRef] [Green Version]
- Houghton, L.C.; Ganmaa, D.; Rosenberg, P.S.; Davaalkham, D.; Stanczyk, F.Z.; Hoover, R.N.; Troisi, R. Associations of Breast Cancer Risk Factors with Premenopausal Sex Hormones in Women with Very Low Breast Cancer Risk. Int. J. Environ. Res. Public Health 2016, 13, 1066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kasi, P.D.; Tamilselvam, R.; Skalicka-Woźniak, K.; Nabavi, S.M.; Daglia, M.; Bishayee, A.; Pazoki-Toroudi, H. Molecular targets of curcumin for cancer therapy: An updated review. Tumor Biol. 2016, 37, 13017–13028. [Google Scholar] [CrossRef] [PubMed]
- Anand, P.; Sundaram, C.; Jhurani, S.; Kunnumakkara, A.B.; Aggarwal, B.B. Curcumin and cancer: An “old-age” disease with an “age-old” solution. Cancer Lett. 2008, 267, 133–164. [Google Scholar] [CrossRef]
- Lai, C.-S.; Ho, C.-T.; Pan, M.-H. The Cancer Chemopreventive and Therapeutic Potential of Tetrahydrocurcumin. Biomolecules 2020, 10, 831. [Google Scholar] [CrossRef] [PubMed]
- Sak, K. Radiosensitizing Potential of Curcumin in Different Cancer Models. Nutr. Cancer 2019, 72, 1276–1289. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhu, D.; Liu, S.; Shao, M.; Liu, Y.; Li, A.; Lv, Y.; Huang, M.; Lou, D.; Fan, Q. Curcumin enhances radiosensitization of nasopharyngeal carcinoma by regulating circRNA network. Mol. Carcinog. 2019, 59, 202–214. [Google Scholar] [CrossRef]
- Wang, Q.; Fan, H.; Liu, Y.; Yin, Z.; Cai, H.; Liu, J.; Wang, Z.; Shao, M.; Sun, X.; Diao, J.; et al. Curcumin enhances the radiosensitivity in nasopharyngeal carcinoma cells involving the reversal of differentially expressed long non-coding RNAs. Int. J. Oncol. 2013, 44, 858–864. [Google Scholar] [CrossRef] [PubMed]
- Marín-Béjar, O.; Mas, A.M.; González, J.; Martinez, D.; Athie, A.; Morales, X.; Galduroz, M.; Raimondi, I.; Grossi, E.; Guo, S.; et al. The human lncRNA LINC-PINT inhibits tumor cell invasion through a highly conserved sequence element. Genome Biol. 2017, 18, 1–15. [Google Scholar] [CrossRef]
- Khan, H.; Reale, M.; Ullah, H.; Sureda, A.; Tejada, S.; Wang, Y.; Zhang, Z.-J.; Xiao, J. Anti-cancer effects of polyphenols via targeting p53 signaling pathway: Updates and future directions. Biotechnol. Adv. 2020, 38, 107385. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zhou, N.; Huang, J.; Liu, Q.; Fukuda, K.; Ma, D.; Lu, Z.; Bai, C.; Watabe, K.; Mo, Y.-Y. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2012, 23, 340–350. [Google Scholar] [CrossRef] [Green Version]
- Kujundžić, R.N.; Grbeša, I.; Ivkić, M.; Katdare, M.; Gall-Trošelj, K. Curcumin downregulates H19 gene transcription in tumor cells. J. Cell. Biochem. 2008, 104, 1781–1792. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, M.; Wang, Y.; Luo, J. Curcumin sensitizes prostate cancer cells to radiation partly via epigenetic activation of miR-143 and miR-143 mediated autophagy inhibition. J. Drug Target. 2017, 25, 645–652. [Google Scholar] [CrossRef]
- Lam, J.S.; Klatte, T.; Breda, A. Staging of renal cell carcinoma: Current concepts. Indian J. Urol. 2009, 25, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Yang, P.; Wang, H.; He, Z.-Y. Silence of long noncoding RNA PANDAR switches low-dose curcumin-induced senescence to apoptosis in colorectal cancer cells. OncoTargets Ther. 2017, 10, 483–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, S.; Mahalanobish, S.; Saha, S.; Ghosh, S.; Sil, P.C. Natural products: An upcoming therapeutic approach to cancer. Food Chem. Toxicol. 2019, 128, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, Y.; Shen, X.; Chen, C.; Ni, H.; Sheng, N.; Hua, M.; Wu, Y. Down-regulation of lncRNA PCGEM1 inhibits cervical carcinoma by modulating the miR-642a-5p/LGMN axis. Exp. Mol. Pathol. 2020, 117, 104561. [Google Scholar] [CrossRef]
- Fu, X.; Ravindranath, L.; Tran, N.; Petrovics, G.; Srivastava, S. Regulation of Apoptosis by a Prostate-Specific and Prostate Cancer-Associated Noncoding Gene, PCGEM1. DNA Cell Biol. 2006, 25, 135–141. [Google Scholar] [CrossRef]
- Negri, A.; Naponelli, V.; Rizzi, F.; Bettuzzi, S. Molecular Targets of Epigallocatechin—Gallate (EGCG): A Special Focus on Signal Transduction and Cancer. Nutrients 2018, 10, 1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, V.; Tuli, H.S.; Tania, M.; Srivastava, S.; Ritzer, E.E.; Pandey, A.; Aggarwal, D.; Barwal, T.S.; Jain, A.; Kaur, G.; et al. Molecular mechanisms of action of epigallocatechin gallate in cancer: Recent trends and advancement. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Singh, B.N.; Shankar, S.; Srivastava, R.K. Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms, perspectives and clinical applications. Biochem. Pharmacol. 2011, 82, 1807–1821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konishi, M.; Imai, A.; Fujii, M.; Sugimoto, K.; Katakami, N.; Imai, Y.; Kamoshida, S. Correlation of Expression Levels of Copper Transporter 1 and Thymidylate Synthase with Treatment Outcomes in Patients with Advanced Non-small Cell Lung Cancer Treated with S-1/Carboplatin Doublet Chemotherapy. Asian Pac. J. Cancer Prev. 2018, 19, 435–441. [Google Scholar] [PubMed]
- Guo, Z.; He, C.; Yang, F.; Qin, L.; Lu, X.; Wu, J. Long non-coding RNA-NEAT1, a sponge for miR-98-5p, promotes expression of oncogene HMGA2 in prostate cancer. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Sui, S.; Wu, H.; Zhang, J.; Zhang, X.; Xu, S.; Pang, D. The transcriptional landscape of lncRNAs reveals the oncogenic function of LINC00511 in ER-negative breast cancer. Cell Death Dis. 2019, 10, 599. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhao, B.; Wang, X.; Zhang, F.; Yu, W. LINC00511 knockdown enhances paclitaxel cytotoxicity in breast cancer via regulating miR-29c/CDK6 axis. Life Sci. 2019, 228, 135–144. [Google Scholar] [CrossRef]
- Hatami, E.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. Gambogic acid: A shining natural compound to nanomedicine for cancer therapeutics. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188381. [Google Scholar] [CrossRef]
- Gan, L.; Yang, Y.; Li, Q.; Feng, Y.; Liu, T.; Guo, W. Epigenetic regulation of cancer progression by EZH2: From biological insights to therapeutic potential. Biomark. Res. 2018, 6, 10. [Google Scholar] [CrossRef]
- Lambrou, G.I.; Hatziagapiou, K.; Zaravinos, A. The Non-Coding RNA GAS5 and Its Role in Tumor Therapy-Induced Resistance. Int. J. Mol. Sci. 2020, 21, 7633. [Google Scholar] [CrossRef]
- Khaitan, D.; Dinger, M.E.; Mazar, J.; Crawford, J.; Smith, M.A.; Mattick, J.S.; Perera, R.J. The Melanoma-Upregulated Long Noncoding RNA SPRY4-IT1 Modulates Apoptosis and Invasion. Cancer Res. 2011, 71, 3852–3862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuli, H.S.; Tuorkey, M.J.; Thakral, F.; Sak, K.; Kumar, M.; Sharma, A.K.; Sharma, U.; Jain, A.; Aggarwal, V.; Bishayee, A. Molecular Mechanisms of Action of Genistein in Cancer: Recent Advances. Front. Pharmacol. 2019, 10, 1336. [Google Scholar] [CrossRef] [Green Version]
- Russo, M.; Russo, G.L.; Daglia, M.; Kasi, P.D.; Ravi, S.; Nabavi, S.M. Understanding genistein in cancer: The “good” and the “bad” effects: A review. Food Chem. 2016, 196, 589–600. [Google Scholar] [CrossRef]
- Lynch, S.M.; O’Neill, K.M.; McKenna, M.M.; Walsh, C.P.; McKenna, D.J. Regulation of miR-200c and miR-141 by Methylation in Prostate Cancer. Prostate 2016, 76, 1146–1159. [Google Scholar] [CrossRef] [PubMed]
- Shao, T.; Hu, Y.; Tang, W.; Shen, H.; Yu, Z.; Gu, J. The long noncoding RNA HOTAIR serves as a microRNA-34a-5p sponge to reduce nucleus pulposus cell apoptosis via a NOTCH1-mediated mechanism. Gene 2019, 715, 144029. [Google Scholar] [CrossRef]
- Mohanan, P.; Subramaniyam, S.; Mathiyalagan, R.; Yang, D.-C. Molecular signaling of ginsenosides Rb1, Rg1, and Rg3 and their mode of actions. J. Ginseng Res. 2018, 42, 123–132. [Google Scholar] [CrossRef]
- Teng, S.; Wang, Y.; Li, P.; Liu, J.; Wei, A.; Wang, H.; Meng, X.; Pan, D.; Zhang, X. Effects of R type and S type ginsenoside Rg3 on DNA methylation in human hepatocarcinoma cells. Mol. Med. Rep. 2017, 15, 2029–2038. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Zhang, J.; Ge, H.; Xu, Y.; Ren, S.; Yue, C.; Li, G.; Wu, J. Long non-coding RNA CASC2 in solid tumors: A meta-analysis. Clin. Chim. Acta 2018, 486, 357–368. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Zhu, G.-Y.; Lu, Y.; Yang, A.; Chen, D.; Huang, H.-J.; Peng, S.-X.; Chen, L.-W.; Li, Y.-W. Luteolin suppresses epithelial-mesenchymal transition and migration of triple-negative breast cancer cells by inhibiting YAP/TAZ activity. Biomed. Pharmacother. 2020, 129, 110462. [Google Scholar] [CrossRef]
- Jiang, C.-Q.; Ma, L.-L.; Lv, Z.-D.; Feng, F.; Chen, Z.; Liu, Z.-D. Polydatin induces apoptosis and autophagy via STAT3 signaling in human osteosarcoma MG-63 cells. J. Nat. Med. 2020, 74, 533–544. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.-H.; Peng, C.; Zhang, H. Polydatin: A review of pharmacology and pharmacokinetics. Pharm. Biol. 2013, 51, 1347–1354. [Google Scholar] [CrossRef]
- Ezzati, M.; Yousefi, B.; Velaei, K.; Safa, A. A review on anti-cancer properties of Quercetin in breast cancer. Life Sci. 2020, 248, 117463. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.-H.; Sethi, G.; Um, J.-Y.; Shanmugam, M.K.; Arfuso, F.; Kumar, A.P.; Bishayee, A.; Ahn, K.S. The Role of Resveratrol in Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 2589. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; He, F.; Chen, L.; Li, Q.; Jin, S.; Zheng, H.; Lin, J.; Zhang, H.; Ma, S.; Mei, J.; et al. Resveratrol inhibits pulmonary fibrosis by regulating miR-21 through MAPK/AP-1 pathways. Biomed. Pharmacother. 2018, 105, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.Q.; Mohamed, E.A.N.; Hakeem, I.; Nazeer, A.; Kuttikrishnan, S.; Prabhu, K.S.; Siveen, K.S.; Nawaz, Z.; Ahmad, A.; Zayed, H.; et al. Sanguinarine Induces Apoptosis in Papillary Thyroid Cancer Cells via Generation of Reactive Oxygen Species. Molecules 2020, 25, 1229. [Google Scholar] [CrossRef] [Green Version]
- Deep, G.; Agarwal, R. Antimetastatic efficacy of silibinin: Molecular mechanisms and therapeutic potential against cancer. Cancer Metastasis Rev. 2010, 29, 447–463. [Google Scholar] [CrossRef] [Green Version]
- Clarke, J.D.; Dashwood, R.H.; Ho, E. Multi-targeted prevention of cancer by sulforaphane. Cancer Lett. 2008, 269, 291–304. [Google Scholar] [CrossRef] [Green Version]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Dashwood, R.H. Epigenetic Regulation of NRF2/KEAP1 by Phytochemicals. Antioxidants 2020, 9, 865. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-Y.; Khor, T.O.; Su, Z.-Y.; Saw, C.L.-L.; Shu, L.; Cheung, K.-L.; Huang, Y.; Yu, S.; Kong, A.-N.T. Epigenetic Modifications of Nrf2 by 3,3′-diindolylmethane In Vitro in TRAMP C1 Cell Line and In Vivo TRAMP Prostate Tumors. AAPS J. 2013, 15, 864–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khor, T.O.; Huang, Y.; Wu, T.-Y.; Shu, L.; Lee, J.; Kong, A.-N.T. Pharmacodynamics of curcumin as DNA hypomethylation agent in restoring the expression of Nrf2 via promoter CpGs demethylation. Biochem. Pharmacol. 2011, 82, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.A.; Piao, M.J.; Hyun, Y.J.; Zhen, A.X.; Cho, S.J.; Ahn, M.J.; Yi, J.M.; Hyun, J.W. Luteolin promotes apoptotic cell death via upregulation of Nrf2 expression by DNA demethylase and the interaction of Nrf2 with p53 in human colon cancer cells. Exp. Mol. Med. 2019, 51, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.-J.; Yu, H.-W.; Yang, Y.-Z.; Wu, W.-Y.; Chen, T.-Y.; Jia, K.-K.; Kang, L.-L.; Jiao, R.-Q.; Kong, L.-D. Polydatin prevents fructose-induced liver inflammation and lipid deposition through increasing miR-200a to regulate Keap1/Nrf2 pathway. Redox Biol. 2018, 18, 124–137. [Google Scholar] [CrossRef]
- Passon, D.M.; Lee, M.; Rackham, O.; Stanley, W.A.; Sadowska, A.; Filipovska, A.; Fox, A.H.; Bond, C.S. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. USA 2012, 109, 4846–4850. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Liu, R.H. Potential Mechanisms of Action of Dietary Phytochemicals for Cancer Prevention by Targeting Cellular Signaling Transduction Pathways. J. Agric. Food Chem. 2018, 66, 3260–3276. [Google Scholar] [CrossRef]
- Hulsen, T.; Jamuar, S.S.; Moody, A.R.; Karnes, J.H.; Varga, O.; Hedensted, S.; Spreafico, R.; Hafler, D.A.; McKinney, E.F. From Big Data to Precision Medicine. Front. Med. 2019, 6, 34. [Google Scholar] [CrossRef] [Green Version]
- Gazdar, A.F. Activating and resistance mutations of EGFR in non-small-cell lung cancer: Role in clinical response to EGFR tyrosine kinase inhibitors. Oncogene 2009, 28 (Suppl. S1), S24–S31. [Google Scholar] [CrossRef] [Green Version]
- Tan, D.S.W.; Chong, F.T.; Leong, H.S.; Toh, S.Y.; Lau, D.P.; Kwang, X.L.; Zhang, X.G.M.; Sundaram, G.M.; San Tan, G.; Chang, M.M.; et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat. Med. 2017, 23, 1167–1175. [Google Scholar] [CrossRef]
- Farhangi, B.; Alizadeh, A.M.; Khodayari, H.; Khodayari, S.; Dehghan, M.J.; Khori, V.; Heidarzadeh, A.; Khaniki, M.; Sadeghiezadeh, M.; Najafi, F. Protective effects of dendrosomal curcumin on an animal metastatic breast tumor. Eur. J. Pharmacol. 2015, 758, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Rizeq, B.; Gupta, I.; Ilesanmi, J.; Alsafran, M.; Rahman, M.; Ouhtit, A. The Power of Phytochemicals Combination in Cancer Chemoprevention. J. Cancer 2020, 11, 4521–4533. [Google Scholar] [CrossRef] [PubMed]
- Bsc, M.Z.A.; Mahomoodally, M.F. Combating breast cancer using combination therapy with 3 phytochemicals: Piperine, sulforaphane, and thymoquinone. Cancer 2019, 125, 1600–1611. [Google Scholar] [CrossRef]
- Ho, J.; Cheung, M. Combination of Phytochemicals as Adjuvants for Cancer Therapy. Recent Pat. Anti-Cancer Drug Discov. 2014, 9, 297–302. [Google Scholar] [CrossRef]
- Subramanian, A.P.; Jaganathan, S.K.; Manikandan, A.; Pandiaraj, K.N.; Gomathi, N.; Supriyanto, E. Recent trends in nano-based drug delivery systems for efficient delivery of phytochemicals in chemotherapy. RSC Adv. 2016, 6, 48294–48314. [Google Scholar] [CrossRef]




| lncRNAs | Type | Related Cancer | Molecular Function | Outcome | Reference |
|---|---|---|---|---|---|
| ANRIL | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation MMP3, caspse-9, caspase-3, Bcl-2, E2F1, c-Myc, and miR-122-5p; Negative correlation p15, p16, TIMP2, Bax, miR-99a, and miR-449a | Induction of cell proliferation, cell cycle, migration, metastasis, and EMT; Inhibition of apoptosis and autophagy cell death | [52,53,54,55] |
| CCAT1/2 | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation GAC | Induction of reprogramming of energy metabolism | [56,57] |
| CRNDE | Oncogenic | Bladder, breast, colon, gastric, leukemia, liver, lung, ovarian, pancreas, renal, SNC, and uterine | Positive correlation GLUT4; Negative correlation Insulin, IGF-I, and IGF-II | Induction of cell proliferation, migration, metastasis, and reprogramming of energy metabolism | [58,59,60] |
| GAS5 | Tumor suppressor | Bladder, breast, colon, gastric, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation PTEN and p21; Negative correlation ERK1/2, NF-κB, CDK6, E2F1, cyclinD1, Vimentin, and MMP2 | Induction of autophagy cell death; Inhibition of cell survival, proliferation, migration, metastasis, and cell cycle | [12,13,17,61] |
| H19 | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation c-Myc, cyclinA2, CDK4, cyclinB1, cyclin D1, and cyclin E1; Negative correlation RB, EGFR, p21, and IGF-II | Induction of cell proliferation, cell cycle, migration, metastasis, EMT, tumor angiogenesis, and immune escape; Inhibition of apoptosis and autophagic cell death | [17,62,63,64] |
| HOTAIR | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation cyclin D1, cyclin E1, CDK4, CDK2, E2F1, P38, Bcl-2, NOTCH1, β-catenin, N-cadherin, Vimentin, Snail, Twist, MMP-9, MMP-2, MMP-3, FGF1, VEGFA, Ang2, and GLUT1; Negative correlation p53, p21, p16, PIK3R3, caspase-9, and caspase-3 | Induction of cell proliferation, cell cycle, invasion, metastasis, EMT, tumor angiogenesis, and immune escape; Inhibition of apoptotic and autophagic cell death | [17,65,66] |
| HULC | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, SNC, and uterine | Positive correlation ZEB1, ZO-1, LC3-II/LC3-I, pmTOR, E2F1, and Snail; Negative correlation E-cadherin | Induction of cell proliferation, migration, and metastasis; Inhibition of apoptotic cell death | [17,18] |
| MALAT1 | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation CDK4, ZEB2, slug, β-catenin, N-cadherin, vimentin, Twist, MMP1, MMP-9, VEGF, TGFA, and TGF-β Negative correlation BAX, E-cadherin, MMP19, TIMP-3, and miR-200s | Induction of cell cycle, cell proliferation, EMT, differentiation, migration, metastasis, chemoresistance and tumor angiogenesis; Inhibition of DNA damage, apoptosis and autophagy cell death | [67,68,69,70,71] |
| MEG3 | Tumor suppressor | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation p53, caspase-3, procaspase-9, cyt. c, and Bax; Negative correlation PI3K, Akt, mTOR cyclin D1, cyclin B1, CDK1, and Bcl-2 | Induction of apoptotic and autophagic cell death; Inhibition of cell proliferation, invasion, metastasis, and cell cycle | [17,72,73,74] |
| PVT1 | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation EZH2, c-Myc, CD115, FGF2, and HIF-1α; Negative correlation miR-31, miR-152, miR-186, and miR-195 | Induction of cell proliferation, migration, metastasis, cell cycle, and angiogenesis; Inhibition of apoptotic cell death | [17,75] |
| TERRA | Tumor suppressor | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Negative correlation TRF2 | Induction of apoptotic cell death; Inhibition of cell proliferation, invasion, metastasis, and cell cycle | [76,77,78] |
| UCA1 | Oncogenic | Bladder, breast, colon, gastric, head and neck, leukemia, liver, lung, osteosarcoma, ovarian, pancreas, prostate, renal, SNC, and uterine | Positive correlation PI3K, AMPK, cyclinD1, ZEB1, ZEB2, N-cadherin, Vimentin, Snail, β-catenin, MMP-7, and FGFR1; Negative correlation p27, E-cadherin, and MMP-14 | Induction of cell proliferation, migration, metastasis, cell cycle, EMT, and reprogramming of energy metabolism; Inhibition of cellular apoptosis | [19,79,80] |
| Phytochemical | Source Plant | Cancer Type | IC50, Exposure Time | Target lncRNAs | Target Genes | Biological Functions | Reference |
|---|---|---|---|---|---|---|---|
| Anacardic acid | Anacardium occidentale L. (Anacardiaceae) | Breast cancer (MCF-7 and MDA-MB-231 cells) | 13.5 µM and 35.0 µM, 6 h | CFLAR-AS1, UBL7-AS1 and MIR210HG↓ | SCD, INSIG1 and TGM2↓PDK4, GPR176 and ZBT20↑ | Inhibits proliferation | [116] |
| Baicalein | Scutellaria baicalensis Georgi | Melanoma (A375 and SK-MEL-28 cells) | 50 µM, 24 h | CCAT1↓ | Wnt-3a, β-catenin, MEK and ERK↓ | Inhibits proliferation, invasion, migration and promotes apoptosis | [117] |
| Cervical cancer (HeLa and SiHa cells) | 100 µM, 24 h | lncRNA- BDLNR↓ | PI3K/Akt pathway↓ | Inhibits tumor growth, migration and proliferation | [23] | ||
| Hepatocellular carcinoma (SMMC-7721, HCCLM3, Hep3B and HepG2 cells) | 12.5 µM, 48 h | NKILA↑ | NF-κB signaling↓ | Inhibits migration, proliferation and promotes apoptosis | [118] | ||
| Breast cancer (MDA-MB-231 and MCF-7 cells) | 50 µM, 48 h | PAX8-AS1-N↑ | miR-17-5p↓ ZBTB4, PTEN and CDKN1A↑ | Inhibits proliferation and cell death | [119] | ||
| Berberis | Coptis chinensis, Berberis poiretii Schneid, Berberis vernae Schneid, Berberis wilsoniae Hemsl | Colorectal cancer (HT29 and HCT116 cells) | 40 µM, 48 h | CASC2↑ | Bcl-2↓ caspase-3 and caspase-9↑ | Inhibits proliferation, viability and promotes apoptosis | [26,120] |
| Non-small cell lung carcinoma (A549 and H1975 cells) | 25 µM, 24 h | HOTAIR↓ | Vimentin↓ E-cadherin and miR-34a-5p↑ | Inhibits proliferation, invasion and metastasis | [27] | ||
| Bharangin | Pygmacopremna herbacea (Roxb), Premna herbacea | Breast cancer (MCF-7, MDAMB-231, MDA-MB-453, MDA-MB-468 and T-47D cells) | 5 µM, 6 h | H19↓ MEG-3, GAS-5, MHRT and NEAT1↑ | NF-κB and Bcl-2↓ Bax↑ | Inhibits proliferation and migration and promotes apoptosis and cell cycle arrest | [121] |
| Calycosin | Radix astragali | Breast cancer (MCF-7, T47D, MDA-MB-468, and SKBR3 cells) | 16 µM, 24 h | WDR7-7↑ | p-SRC, p-EGFR, p-ERK1/2, p-Akt and GPR30↓ | Inhibits proliferation and tumor growth | [122] |
| Breast cancer (MCF-7 cells) | 80 µM, 48 h | HOTAIR↓ | p-Akt↓ | Inhibits proliferation and promotes apoptosis | [123] | ||
| Nasopharyngeal carcinoma (CNE1, CNE2 and C666-1 cells) | 50 µM, 48 h | EWSAT1↓ | p- IκBα, p-c-Jun, TRAF6 and p-TAK1↓ | Inhibits proliferation and tumor growth | [124] | ||
| Curcumin | Curcuma longa L. | Pancreatic ductal adenocarcinoma (BxPC3-GemR and Panc1 cells) | 8 µM and 20 µM, 48 h | lncRNA-PVT1↓ | EZH2 and SUZ12↓ | Promotes the sensitivity of BxPC3-GemR cells to gemcitabine | [125] |
| Acute lymphoblastic leukemia (LBH-589 cell) | 30 µM, 48 h | linc-PINT↑ | HOMX1↑ | Promotes apoptosis and cell cycle arrest | [126] | ||
| Gastric cancer (GES-1 cells) | 50 µM, 48 h | H19↓ | Myc, Bcl-2↓ p53 and Bax↑ | Promotes apoptosis | [24] | ||
| Prostate cancer stem cells (Du145 and 22RV cells) | 46.5 µM, 48 h | lncRNA-ROR↓ | Oct4, CDK4 and cyclin D1↓ miR-145↑ | Inhibits invasion, proliferation, xenograft growth and cell cycle (G2/M) | [127] | ||
| Renal cell carcinoma | 20 µM, 24 h | HOTAIR↓ XIST↑ | miR-106b↓ p21↑ | Inhibits migration, invasion and proliferation | [128,129] | ||
| Breast cancer (MCF7, MDA-MB231 and SKBR3 cells) | 13.5 µM, 48 h | Tusc7 and GAS5↑ | - | Inhibits proliferation and promotes apoptosis and cell cycle | [130] | ||
| Colorectal carcinoma (HCT116 and SW480 cells) | 10 µM, 24 h | lncRNA-NBR2↑ | AMPK/mTOR signaling axis↓ | Inhibits proliferation | [131] | ||
| Hepatocellular cancer (HepG2 and HuH-7 cells) | 23 µM, 48 h | MEG3↑ | DNMT1, DNMT3A DNMT3B↓ miR-29a and miR-185↑ | Inhibits proliferation | [132] | ||
| Ovarian cancer (OVCAR-3 and SKOV3 cells) | 1 µM, 36 h | MEG3↑ | miR-214↓ | Inhibits cisplatin resistance | [133] | ||
| Lung cancer (A549 cells) | 1 µM, 24 h | lncRNA-UCA1↓ | Cyclin D1 and Wnt/mTOR signaling↓ caspase-3 and caspase-9↑ | Inhibits proliferation and promotes apoptosis | [134] | ||
| DIM | Brassica vegetables, Brussels sprouts, Cruciferous vegetables | Prostate cancer (LNCaP and CWR22Rv1 cells) | 20 µM, 48 h | PCGEM1↓ | p54/nrb↓ | Inhibits castration resistance | [135] |
| Colon cancer (HT-29 and HCT-116 cells) | 30 µM, 72 h | HOTAIR, CCAT1-L↓ | - | - | [136] | ||
| EGCG | Camellia sinensis (L.) Kuntze (Theaceae) | Non-small cell lung carcinoma (A549 cells and H460 cells) | 20 µM, 24 h | lncRNA-NEAT↑ | p-ERK1/2 and miR-98-5p↓ | Promotes cisplatin sensitivity | [137] |
| Osteosarcoma (U2OS and SaoS2 cells) | 20 μg/mL, 24 h | SOX2OT V7↓ | Notch3/DLL3 signaling↓ | Inhibits stemness and autophagy and promotes death | [138] | ||
| Gastric cancer (AGS and SGC7901 cells) | 100 μM, 48 h | LINC00511↓ | miR-29b↑ | Inhibits invasion, proliferation, migration and gemcitabine resistance | [139] | ||
| Gambogic acid | Garcinia hurburyi | Bladder cancer (EJ, UMUC3 and T24T cells) | 1 μM, 48 h | SPRY4-IT1↓ | EZH2↓ miR-101↑ | Inhibits proliferation, migration and invasion, and promotes apoptosis | [140] |
| Bladder cancer (T24 and EJ cells) | 2 μM, 48 h | GAS5↑ | EZH2↓ | Inhibits viability, xenograft growth and promotes apoptosis | [141] | ||
| Genistein | Genista tinctoria L., Glycine max L. | Breast cancer (MCF-7 cells) | 80 μM, 48 h | HOTAIR↓ | p-Akt↓ | Inhibits proliferation and promotes apoptosis | [123] |
| Renal carcinoma, colorectal adenocarcinoma and prostate cancer | 25 μM, 96 h | HOTAIR↓ | miR-141↑ | Inhibits proliferation and promotes apoptosis | [142] | ||
| Prostate cancer (PC3 and DU145 PCa cells) | 25 μM, 96 h | HOTAIR↓ | miR-34a↑ | Inhibits proliferation, migration and invasion | [143] | ||
| Colorectal cancer (SW480 cells) | 50 μM, 48 h | lncRNA- TTTY18↓ | Akt, SGK1 and p38 MAPK↓ | Inhibits proliferation and migration | [144] | ||
| Ginsenosides | Panax ginseng, P. quinquefoliu, P. vietnamensis, P. japonicas, P. notoginseng | Breast cancer (MCF-7 cells) | 30 μM, 24 h | C3orf67-AS1↓ | - | Inhibits colonization and proliferation and promotes apoptosis | [145] |
| Breast cancer (MCF-7 cells) | 20 μM, 24 h | RFX3-AS1↓ STXBP5-AS1↑ | RFX3, SLC1A1, PUM3 and STXbp5↓ GRM1↑ | Inhibits colonization | [146] | ||
| Pancreatic cancer (Panc-1 and SW1990 cells) | 50 μM, 24 h | CASC2↑ | PTEN signaling↑ | Inhibits proliferation and tumor growth | [147] | ||
| Colorectal cancer (Caco-2 cells) | 50 µM, 24 h | CCAT1↓ | Bcl-2, vimentin and CCND1↓ Bax, p53 and caspase-3↑ | Inhibits proliferation, invasion and migration | [148] | ||
| Breast cancer (MCF-7 cells) | 80 µM and 40 µM, 24 h | lncRNA-H19↓ | PKM2↓ | Inhibits tumorigenesis and the Warburg effect | [149] | ||
| Hyperoside | Artemisia capillaris, Apocynum venetum | Non-small-cell lung carcinoma (NCI-H1975 cells) | 87.4 µM, 48 h | CCAT1↓ | FoxO1↑ | Inhibits proliferation, xenografts growth and promotes apoptosis | [150] |
| Luteolin | Passiflora edulis,Taraxacum officinale | Thyroid carcinoma (IHH-4, FTC-133 and 8505C cells) | 10 µM, 24 h | BANCR↓ | CCND1, p-CREB, PCNA and TSHR signaling↓ | Inhibits cell cycle, proliferation and xenograft growth | [151] |
| Polydatin | Polygonum cuspidatum Sieb. et Zucc. (Polygonaceae) Fallopia japonica (Houtt.) | Osteosarcoma (Saos-2 and MG-63 cells) | 150 µM, 48 h | TUG1↓ | p-Akt↓ | Inhibits proliferation, tumor volume and tumor weight and promotes apoptosis | [152] |
| Quercetin | Allium cepa L. (Amaryllidaceae) | Prostate cancer (PC-3 cells) | 50 µM, 48 h | MALAT1↓ | N-cadherin, p-Akt and Bcl-2↓ E-cadherin and Bax↑ | Inhibits proliferation, migration, invasion, xenografts growth and EMT and promotes apoptosis | [153] |
| Resveratrol | Grapes, blueberries, Morus alba L., Polygonum cuspidatum Sieb. et Zucc., Rubus idaeus L. | Prostate cancer (DU145 and LNCaP cells) | 25 µM, 24 h | PCAT29↑ | IL-6, STAT3 and miR-21↓ PDCD4↑ | Inhibits proliferation and tumorigenesis | [154] |
| Multiple myeloma (U266 and LP1 cells) | 40 µM, 72 h | NEAT1↓ | Survivin, β-catenin, MMP-7 and c-Myc↓ | Inhibits migration, proliferation and invasion | [155] | ||
| Colorectal cancer (LoVo and HCT116 cells) | 50 µM, 48 h | MALAT1↓ | c-Myc, MMP-7 and β-catenin↓ | Inhibits migration, proliferation and invasion | [156] | ||
| Breast cancer (LTED and MCF-7 cells) | 50 µM, 24 h | u-Eleanor↓ | ER gene↓ | Inhibits cell growth | [157] | ||
| Lung cancer (A549 cells) | 40 µM, 48 h | AK001796↓ | - | Promotes cell cycle arrest (G0/G1) and apoptosis | [158] | ||
| Sanguinarine | Sanguinaria canadensis L. (bloodroot) | Ovarian cancer (SKOV3 cells) | 5 µM, 48 h | CASC2↑ | NF-κB, PI3K, p-Akt↓ | Inhibits viability, migration and invasion and promotes cell apoptosis | [159] |
| Silibinin | Silybum marianum (L.) Gaertn. (Asteraceae) | Bladder cancer (T24 and UM-UC-3 cells) | 10 µM, 48 h | HOTAIR and ZFAS1↓ | EGFR, SOS1, Ras, PAK1, DDR1, H3K4 and p-Akt↓ | Inhibits proliferation, migration and invasion and promotes apoptosis | [160] |
| Sulforaphane | Brassica oleracea | Prostate cancer (PC-3 and LNCaP cells) | 15 µM, 24 h | LINC01116↓ | MAP1LC3B2 and H2AFY↑ | Inhibits proliferation and clonogenic survival | [161] |
| Colon cancer (HCT116 and HT29 cells) | 15 µM, 24 h | Loc344887↑ (NMRAL2P) | - | Inhibits proliferation colony formation and migration | [162] |
| Phytochemical | Cancer Type | Dose | Target lncRNAs | Biological Functions | Reference |
|---|---|---|---|---|---|
| Baicalein | Cervical cancer | 10 mg/kg/day | lncRNA- BDLNR↓ | Inhibits tumor growth | [23] |
| Hepatocellular carcinoma | 10 mg/kg/day | NKILA↑ | Inhibits tumor growth | [118] | |
| Breast cancer | 10 mg/kg/day | PAX8-AS1-N↑ | Inhibits tumor growth | [119] | |
| Berberis | Non-small cell lung carcinoma | 25 mg/kg/day | HOTAIR↓ | Inhibits tumor growth | [27] |
| Calycosin | Breast cancer | 55 mg/kg/day | WDR7-7↑ | Inhibits tumor growth | [122] |
| Nasopharyngeal carcinoma | 60 mg/kg/day | EWSAT1↓ | Inhibits tumor growth | [124] | |
| Curcumin | Pancreatic ductal adenocarcinoma | 100 mg/kg/day | lncRNA-PVT1↓ | Inhibits tumor growth | [125] |
| DIM | Prostate cancer | 20 mg/kg/day | PCGEM1↓ | Inhibits tumor growth | [135] |
| EGCG | Non-small cell lung carcinoma | 20 mg/kg/day | lncRNA-NEAT1↑ | Inhibits tumor growth | [137] |
| Osteosarcoma | 30 mg/kg/day | SOX2OT V7↓ | Inhibits tumor growth | [138] | |
| Genistein | Colorectal cancer | 30 mg/kg/day | lncRNA-TTTY18↓ | Inhibits tumor growth | [144] |
| Ginsenosides | Pancreatic cancer | 40 mg/kg/day | CASC2↑ | Inhibits tumor growth | [147] |
| Hyperoside | Non-small-cell lung carcinoma | 25 mg/kg/day | CCAT1↓ | Inhibits tumor growth | [150] |
| Luteolin | Thyroid carcinoma | 50 mg/kg/day | BANCR↓ | Inhibits tumor growth | [151] |
| Polydatin | Osteosarcoma | 150 mg/kg/day | TUG1↓ | Inhibits tumor growth | [152] |
| Quercetin | Prostate cancer | 75 mg/kg/day | MALAT1↓ | Inhibits tumor growth | [153] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kalhori, M.R.; Khodayari, H.; Khodayari, S.; Vesovic, M.; Jackson, G.; Farzaei, M.H.; Bishayee, A. Regulation of Long Non-Coding RNAs by Plant Secondary Metabolites: A Novel Anticancer Therapeutic Approach. Cancers 2021, 13, 1274. https://doi.org/10.3390/cancers13061274
Kalhori MR, Khodayari H, Khodayari S, Vesovic M, Jackson G, Farzaei MH, Bishayee A. Regulation of Long Non-Coding RNAs by Plant Secondary Metabolites: A Novel Anticancer Therapeutic Approach. Cancers. 2021; 13(6):1274. https://doi.org/10.3390/cancers13061274
Chicago/Turabian StyleKalhori, Mohammad Reza, Hamid Khodayari, Saeed Khodayari, Miko Vesovic, Gloria Jackson, Mohammad Hosein Farzaei, and Anupam Bishayee. 2021. "Regulation of Long Non-Coding RNAs by Plant Secondary Metabolites: A Novel Anticancer Therapeutic Approach" Cancers 13, no. 6: 1274. https://doi.org/10.3390/cancers13061274