LncRNA PART1 Promotes Proliferation and Migration, Is Associated with Cancer Stem Cells, and Alters the miRNA Landscape in Triple-Negative Breast Cancer
Abstract
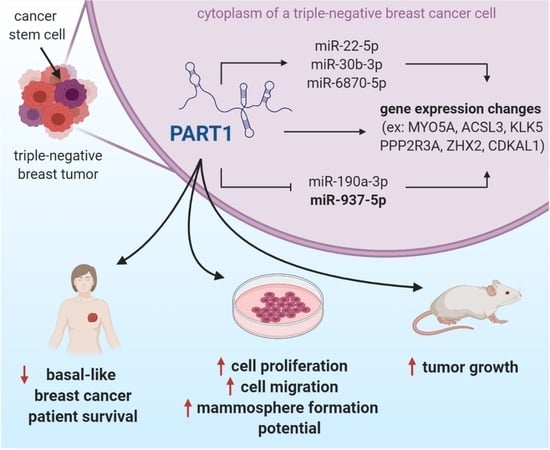
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line Models and Tumor Studies
2.2. R1881 and D36 Treatment in Cell Culture
2.3. Total RNA Extraction and Reverse Transcription-Quantitative PCR
2.4. Knockdown of PART1, Cell Proliferation, Migration, Apoptosis, Mammosphere Assays
2.5. Aldefluor Analysis and Sorting
2.6. Western Blotting
2.7. Subcellular Localization of PART1
2.8. Dataset Analyses
2.9. Transcriptome Analysis
2.10. MiRNA Analyses
2.11. Luciferase Reporter Assay for PART1—miR-937-5p Interaction
2.12. Statistics
3. Results
3.1. PART1 Is Enriched in Basal-Like/Triple-Negative Breast Cancer Patient Tumors and Is Androgen Inducible in Breast Cancer Cells
3.2. PART1 Is Oncogenic in Triple-Negative Breast Cancer Cells
3.3. PART1 Expression Is Associated with Stemness Gene Expression, Aldefluorhigh Cancer Stem Cells, and Contributes to Mammosphere Formation Potential
3.4. PART1 Induces Gene Expression Changes in Triple-Negative Breast Cancer Cells and Is Predominately Cytoplasmic
3.5. PART1 Alters the miRNA Landscape in Triple-Negative Breast Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandya, G.; Kirtonia, A.; Sethi, G.; Pandey, A.K.; Garg, M. The implication of long non-coding RNAs in the diagnosis, pathogenesis and drug resistance of pancreatic ductal adenocarcinoma and their possible therapeutic potential. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188423. [Google Scholar] [CrossRef] [PubMed]
- Kansara, S.; Pandey, V.; Lobie, P.E.; Sethi, G.; Garg, M.; Pandey, A.K. Mechanistic Involvement of Long Non-Coding RNAs in Oncotherapeutics Resistance in Triple-Negative Breast Cancer. Cells 2020, 9, 1511. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prensner, J.R.; Chinnaiyan, A.M. The Emergence of lncRNAs in Cancer Biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidovic, D.; Huynh, T.T.; Konda, P.; Dean, C.; Cruickshank, B.M.; Sultan, M.; Coyle, K.M.; Gujar, S.; Marcato, P. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2019. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wu, J.; Li, X.; Wang, C.; Wang, M.; Liu, J.; Yang, G. NEAT1/miR-124/STAT3 feedback loop promotes breast cancer progression. Int. J. Oncol. 2019. [Google Scholar] [CrossRef]
- Arun, G.; Spector, D.L. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019, 16, 860–863. [Google Scholar] [CrossRef]
- Zhang, Y.; He, Q.; Hu, Z.; Feng, Y.; Fan, L.; Tang, Z.; Yuan, J.; Shan, W.; Li, C.; Hu, X.; et al. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat. Struct. Mol. Biol. 2016, 23, 522–530. [Google Scholar] [CrossRef]
- Peddi, P.F.; Ellis, M.J.; Ma, C. Molecular basis of triple negative breast cancer and implications for therapy. Int. J. Breast Cancer 2012, 2012, 217185. [Google Scholar] [CrossRef]
- Shipitsin, M.; Campbell, L.L.; Argani, P.; Weremowicz, S.; Bloushtain-Qimron, N.; Yao, J.; Nikolskaya, T.; Serebryiskaya, T.; Beroukhim, R.; Hu, M.; et al. Molecular definition of breast tumor heterogeneity. Cancer Cell 2007, 11, 259–273. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ma, F.; Wang, H.; Lin, C.; Fan, Y.; Zhang, X.; Qian, H.; Xu, B. Stem cell marker aldehyde dehydrogenase 1 (ALDH1)-expressing cells are enriched in triple-negative breast cancer. Int. J. Biol. Markers 2013, 28, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Giatromanolaki, A.; Sivridis, E.; Fiska, A.; Koukourakis, M.I. The CD44+/CD24- phenotype relates to “triple-negative” state and unfavorable prognosis in breast cancer patients. Med. Oncol. 2011, 28, 745–752. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Sarkissyan, M.; Elshimali, Y.; Vadgama, J.V. Triple Negative Breast Tumors in African-American and Hispanic/Latina Women Are High in CD44+, Low in CD24+, and Have Loss of PTEN. PLoS ONE 2013, 8, e78259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsang, J.Y.S.; Huang, Y.-H.; Luo, M.-H.; Ni, Y.-B.; Chan, S.-K.; Lui, P.C.W.; Yu, A.M.C.; Tan, P.H.; Tse, G.M. Cancer stem cell markers are associated with adverse biomarker profiles and molecular subtypes of breast cancer. Breast Cancer Res. Treat. 2012, 136, 407–417. [Google Scholar] [CrossRef] [PubMed]
- Perrone, G.; Gaeta, L.M.; Zagami, M.; Nasorri, F.; Coppola, R.; Borzomati, D.; Bartolozzi, F.; Altomare, V.; Trodella, L.; Tonini, G.; et al. In Situ Identification of CD44+/CD24− Cancer Cells in Primary Human Breast Carcinomas. PLoS ONE 2012, 7, e43110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ricardo, S.; Vieira, A.F.; Gerhard, R.; Leitão, D.; Pinto, R.; Cameselle-Teijeiro, J.F.; Milanezi, F.; Schmitt, F.; Paredes, J.; Leitao, D.; et al. Breast cancer stem cell markers CD44, CD24 and ALDH1: Expression distribution within intrinsic molecular subtype. J. Clin. Pathol. 2011, 64, 937–946. [Google Scholar] [CrossRef]
- Idowu, M.O.; Kmieciak, M.; Dumur, C.; Burton, R.S.; Grimes, M.M.; Powers, C.N.; Manjili, M.H. CD44+/CD24−/lowcancer stem/progenitor cells are more abundant in triple-negative invasive breast carcinoma phenotype and are associated with poor outcome. Hum. Pathol. 2012, 43, 364–373. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lennox, K.A.; Behlke, M.A. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016, 44, 863–877. [Google Scholar] [CrossRef] [Green Version]
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The Noncoding RNA MALAT1 Is a Critical Regulator of the Metastasis Phenotype of Lung Cancer Cells. Cancer Res. 2013, 73, 1180–1189. [Google Scholar] [CrossRef] [Green Version]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef]
- Schoch, K.M.; Miller, T.M. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases. Neuron 2017, 94, 1056–1070. [Google Scholar] [CrossRef] [Green Version]
- Lin, B.; White, J.T.; Ferguson, C.; Bumgarner, R.; Friedman, C.; Trask, B.; Ellis, W.; Lange, P.; Hood, L.; Nelson, P.S. PART-1: A novel human prostate-specific, androgen-regulated gene that maps to chromosome 5q12. Cancer Res. 2000, 60, 858–863. [Google Scholar] [PubMed]
- Sun, M.; Geng, D.; Li, S.; Chen, Z.; Zhao, W. LncRNA PART1 modulates toll-like receptor pathways to influence cell proliferation and apoptosis in prostate cancer cells. Biol. Chem. 2018, 399, 387–395. [Google Scholar] [CrossRef]
- Li, M.; Zhang, W.; Zhang, S.; Wang, C.; Lin, Y. PART1 expression is associated with poor prognosis and tumor recurrence in stage I-III non-small cell lung cancer. J. Cancer 2017, 8, 1795–1800. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Ren, J.; Zheng, X.; Hu, X.; Hu, M. Comprehensive analysis of expression profiles of long non-coding RNAs with associated ceRNA network involved in gastric cancer progression. Mol. Med. Rep. 2019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Ma, Z.; He, Y.; Liu, W.; Su, Y.; Tang, Z. PART-1 functions as a competitive endogenous RNA for promoting tumor progression by sponging miR-143 in colorectal cancer. Biochem. Biophys. Res. Commun. 2017, 490, 317–323. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Ren, M.; Li, Y.; Fu, Y.; Deng, M.; Li, C. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J. Exp. Clin. Cancer Res. 2018, 37, 171. [Google Scholar] [CrossRef]
- Zhang, X.-Q.; Sun, S.; Lam, K.-F.; Kiang, K.M.-Y.; Pu, J.K.-S.; Ho, A.S.-W.; Lui, W.-M.; Fung, C.-F.; Wong, T.S.; Leung, G.K.K. A long non-coding RNA signature in glioblastoma multiforme predicts survival. Neurobiol. Dis. 2013, 58, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Piao, L.; Sun, G.; Lv, C.; Jing, Y.; Jin, R. Long Non-Coding RNA PART1 Exerts Tumor Suppressive Functions in Glioma via Sponging miR-190a-3p and Inactivation of PTEN/AKT Pathway. OncoTargets Ther. 2020, 13, 1073–1086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Xu, R. lncRNA PART1 Promotes Breast Cancer Cell Progression by Directly Targeting miR-4516. Cancer Manag. Res. 2020, 12, 7753–7760. [Google Scholar] [CrossRef]
- Lombardo, Y.; de Giorgio, A.; Coombes, C.R.; Stebbing, J.; Castellano, L. Mammosphere Formation Assay from Human Breast Cancer Tissues and Cell Lines. J. Vis. Exp. 2015, 97, e52671. [Google Scholar] [CrossRef] [Green Version]
- Alam, M.; Rajabi, H.; Ahmad, R.; Jin, C.; Kufe, D. Targeting the MUC1-C oncoprotein inhibits self-renewal capacity of breast cancer cells. Oncotarget 2014, 5, 2622–2634. [Google Scholar] [CrossRef] [Green Version]
- Marcato, P.; Dean, C.A.; Pan, D.; Araslanova, R.; Gillis, M.; Joshi, M.; Helyer, L.; Pan, L.; Leidal, A.; Gujar, S.; et al. Aldehyde Dehydrogenase Activity of Breast Cancer Stem Cells Is Primarily Due to Isoform ALDH1A3 and Its Expression Is Predictive of Metastasis. Stem Cells 2011, 29, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Sultan, M.; Vidovic, D.; Paine, A.S.; Huynh, T.T.; Coyle, K.M.; Thomas, M.L.; Cruickshank, B.M.; Dean, C.A.; Clements, D.R.; Kim, Y.; et al. Epigenetic Silencing of TAP1 in Aldefluor+Breast Cancer Stem Cells Contributes to Their Enhanced Immune Evasion. Stem Cells 2018, 36, 641–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mas-Ponte, D.; Carlevaro-Fita, J.; Palumbo, E.; Pulido, T.H.; Guigo, R.; Johnson, R. LncATLAS database for subcellular localization of long noncoding RNAs. RNA 2017, 23, 1080–1087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashid, F.; Shah, A.; Shan, G. Long Non-coding RNAs in the Cytoplasm. Genom. Proteom. Bioinform. 2016, 14, 73–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2009, 123, 725–731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broad Institute. Cancer Cell Line Encyclopedia (CCLE). Available online: https://portals.broadinstitute.org/ccle (accessed on 12 May 2021).
- Paraskevopoulou, M.D.; Vlachos, I.; Karagkouni, D.; Georgakilas, G.; Kanellos, I.; Vergoulis, T.; Zagganas, K.; Tsanakas, P.; Floros, E.; Dalamagas, T.; et al. DIANA-LncBase v2: Indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016, 44, D231–D238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef]
- Sabatier, R.; Finetti, P.; Guille, A.; Adélaïde, J.; Chaffanet, M.; Viens, P.; Birnbaum, D.; Bertucci, F. Claudin-low breast cancers: Clinical, pathological, molecular and prognostic characterization. Mol. Cancer 2014, 13, 228. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.L.; De Antueno, R.; Coyle, K.M.; Sultan, M.; Cruickshank, B.M.; Giacomantonio, M.A.; Giacomantonio, C.A.; Duncan, R.; Marcato, P. Citral reduces breast tumor growth by inhibiting the cancer stem cell marker ALDH1A3. Mol. Oncol. 2016, 10, 1485–1496. [Google Scholar] [CrossRef] [Green Version]
- Zhao, R.; Peng, C.; Song, C.; Zhao, Q.; Rong, J.; Wang, H.; Ding, W.; Wang, F.; Xie, Y. BICC1 as a novel prognostic biomarker in gastric cancer correlating with immune infiltrates. Int. Immunopharmacol. 2020, 87, 106828. [Google Scholar] [CrossRef]
- Alves, C.; Moraes, M.H.; Sousa, J.F.; Pontes, C.L.S.; Ramão, A.; Yokoyama, S.; Trindade, D.M.; Fisher, D.E.; Espreafico, E.M. Myosin-Va Contributes to Manifestation of Malignant-Related Properties in Melanoma Cells. J. Investig. Dermatol. 2013, 133, 2809–2812. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Zhang, W.; Ji, W. MYO5A inhibition by miR-145 acts as a predictive marker of occult neck lymph node metastasis in human laryngeal squamous cell carcinoma. OncoTargets Ther. 2018, 11, 3619–3635. [Google Scholar] [CrossRef] [Green Version]
- Jin, X.; Yan, Y.; Wang, D. ZHX2 is an oncogenic driver of kidney cancer. Cancer Discov. 2018, 8, 1057. [Google Scholar]
- Dmitriev, A.A.; Beniaminov, A.D.; Melnikova, N.V.; Pushkova, E.N.; Gerashchenko, G.V.; Kudryavtseva, A.V.; Kashuba, V.I. Functional Hypermethylation of ALDH1L1, PLCL2, and PPP2R3A in Colon Cancer. Mol. Biol. 2020, 54, 204–211. [Google Scholar] [CrossRef]
- Zhu, D.; Yu, Y.; Wang, W.; Wu, K.; Liu, D.; Yang, Y.; Zhang, C.; Qi, Y.; Zhao, S. Long noncoding RNA PART1 promotes progression of non-small cell lung cancer cells via JAK-STAT signaling pathway. Cancer Med. 2019, 8, 6064–6081. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.-J.; Jiang, D.-M. LncRNA PART1 modulates chondrocyte proliferation, apoptosis, and extracellular matrix degradation in osteoarthritis via regulating miR-373-3p/SOX4 axis. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8175–8185. [Google Scholar] [CrossRef]
- Lou, T.; Ke, K.; Zhang, L.; Miao, C.; Liu, Y. LncRNA PART1 facilitates the malignant progression of colorectal cancer via miR-150-5p/LRG1 axis. J. Cell. Biochem. 2020, 121, 4271–4281. [Google Scholar] [CrossRef]
- Zhou, T.; Wu, L.; Ma, N.; Tang, F.; Zong, Z.; Chen, S. LncRNA PART1 regulates colorectal cancer via targeting miR-150-5p/miR-520h/CTNNB1 and activating Wnt/β-catenin pathway. Int. J. Biochem. Cell Biol. 2020, 118, 105637. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Venkatesh, J.; Wasson, M.-C.D.; Brown, J.M.; Fernando, W.; Marcato, P. LncRNA-miRNA axes in breast cancer: Novel points of interaction for strategic attack. Cancer Lett. 2021, 509, 81–88. [Google Scholar] [CrossRef]
- Clément, T.; Salone, V.; Rederstorff, M. Dual Luciferase Gene Reporter Assays to Study miRNA Function. Methods Mol. Biol. 2015, 1296, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y. Sequence requirements for micro RNA processing and function in human cells. RNA 2003, 9, 112–123. [Google Scholar] [CrossRef] [Green Version]
- Ni, W.J.; Leng, X.M. Dynamic miRNA-mRNA paradigms: New faces of miRNAs. Biochem. Biophys. Rep. 2015, 4, 337–341. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; He, Y.; Yang, S.; Hu, J.; Zhang, Q.; Chen, W.; Xu, H.; Zhang, H.; Zhong, S.; Zhao, J.; et al. The regulatory roles of lncRNAs in the process of breast cancer invasion and metastasis. Biosci. Rep. 2018, 38, BSR20180772. [Google Scholar] [CrossRef] [Green Version]
- Walsh, H.R.; Cruickshank, B.M.; Brown, J.M.; Marcato, P. The Flick of a Switch: Conferring Survival Advantage to Breast Cancer Stem Cells Through Metabolic Plasticity. Front. Oncol. 2019, 9, 753. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 20. [Google Scholar] [CrossRef] [Green Version]
- Dunagin, M.; Cabili, M.N.; Rinn, J.; Raj, A. Visualization of lncRNA by Single-Molecule Fluorescence In Situ Hybridization. Methods Mol. Biol. 2014, 1262, 3–19. [Google Scholar] [CrossRef]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol. 2010, 6, 363. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Han, H.; Zuo, H.; Chen, Z.; Du, Y.; Zhao, W.; Gu, J.; Zhang, Z. Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. Int. J. Cancer 2009, 126, 53–64. [Google Scholar] [CrossRef]
- Wang, H.; Guo, Y.; Mi, N.; Zhou, L. miR-101-3p and miR-199b-5p promote cell apoptosis in oral cancer by targeting BICC1. Mol. Cell. Probes 2020, 52, 101567. [Google Scholar] [CrossRef]
- Dunwell, T.L.; Hesson, L.B.; Pavlova, T.V.; Zabarovska, V.; Kashuba, V.I.; Catchpoole, D.; Chiaramonte, R.; Brini, A.T.; Griffiths, M.; Maher, E.R.; et al. Epigenetic analysis of childhood acute lymphoblastic leukemia. Epigenetics 2009, 4, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Liu, F.; Ma, H.; Cui, X.; Yang, S.; Qin, R. circCDYL Acts as a Tumor Suppressor in Triple Negative Breast Cancer by Sponging miR-190a-3p and Upregulating TP53INP1. Clin. Breast Cancer 2020, 20, 422–430. [Google Scholar] [CrossRef]
- Kumar, B.; Khaleghzadegan, S.; Mears, B.; Hatano, K.; Kudrolli, T.A.; Chowdhury, W.H.; Yeater, D.B.; Ewing, C.M.; Luo, J.; Isaacs, W.B.; et al. Identification of miR-30b-3p and miR-30d-5p as direct regulators of androgen receptor signaling in prostate cancer by complementary functional microRNA library screening. Oncotarget 2016, 7, 72593–72607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, M.; Wang, S.; Fritah, S.; Wang, X.; Zhou, W.; Yang, N.; Ni, S.; Huang, B.; Chen, A.; Li, G.; et al. Interfering with long non-coding RNA MIR22HG processing inhibits glioblastoma progression through suppression of Wnt/β-catenin signalling. Brain 2020, 143, 512–530. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Xu, W.; Gong, J.; Wang, L.; Dai, M.; Chen, G.; Yuan, L. miR-937-5p targets SOX17 to modulate breast cancer cell cycle and cell proliferation through the Wnt signaling pathway. Cell. Signal. 2021, 77, 109818. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, Y.; Li, W.; Zhu, J. CircFNDC3B sequestrates miR-937-5p to derepress TIMP3 and inhibit colorectal cancer progression. Mol. Oncol. 2020, 14, 2960–2984. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruickshank, B.M.; Wasson, M.-C.D.; Brown, J.M.; Fernando, W.; Venkatesh, J.; Walker, O.L.; Morales-Quintanilla, F.; Dahn, M.L.; Vidovic, D.; Dean, C.A.; et al. LncRNA PART1 Promotes Proliferation and Migration, Is Associated with Cancer Stem Cells, and Alters the miRNA Landscape in Triple-Negative Breast Cancer. Cancers 2021, 13, 2644. https://doi.org/10.3390/cancers13112644
Cruickshank BM, Wasson M-CD, Brown JM, Fernando W, Venkatesh J, Walker OL, Morales-Quintanilla F, Dahn ML, Vidovic D, Dean CA, et al. LncRNA PART1 Promotes Proliferation and Migration, Is Associated with Cancer Stem Cells, and Alters the miRNA Landscape in Triple-Negative Breast Cancer. Cancers. 2021; 13(11):2644. https://doi.org/10.3390/cancers13112644
Chicago/Turabian StyleCruickshank, Brianne M., Marie-Claire D. Wasson, Justin M. Brown, Wasundara Fernando, Jaganathan Venkatesh, Olivia L. Walker, Fiorella Morales-Quintanilla, Margaret L. Dahn, Dejan Vidovic, Cheryl A. Dean, and et al. 2021. "LncRNA PART1 Promotes Proliferation and Migration, Is Associated with Cancer Stem Cells, and Alters the miRNA Landscape in Triple-Negative Breast Cancer" Cancers 13, no. 11: 2644. https://doi.org/10.3390/cancers13112644