Getting Lost in the Cell–Lysosomal Entrapment of Chemotherapeutics
Abstract
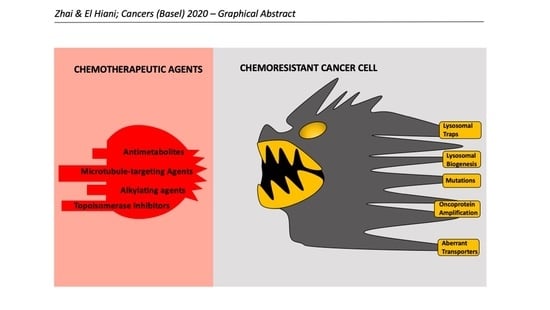
:Simple Summary
Abstract
1. History of Chemotherapy
2. Conventional Chemotherapeutic Compounds and Cell Death
2.1. Antimetabolites
2.2. Alkylating Agents
2.3. Topoisomerase Inhibitors
2.4. Microtubule-Targeting Agents (Vinca Alkaloids and Taxanes)
2.5. Cell-Fate Determination Post-Chemotherapeutic Challenge through Lysosomes
3. Lysosome-Mediated Acquired Chemoresistance
3.1. Organellar Physiology of Lysosomes
3.2. The Endocytic Origin and Metabolic Signaling of Lysosomes
3.3. Lysosomal Abberations in Cancer and Chemoresistance
3.4. Lysosomal Entrapment of Weak-Base Compounds
3.5. Novel Therapeutic Strategies for Lysosome-Mediated Chemoresistance
4. Concluding Remarks
Funding
Conflicts of Interest
References
- Kaufmann, S.H. Paul Ehrlich: Founder of chemotherapy. Nat. Rev. Drug Discov. 2008, 7, 373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krumbhaar, E.B. Rôle of the blood and the bone marrow in certain forms of gas poisoning: I. peripheral blood changes and their significance. JAMA 1919, 72, 39–41. [Google Scholar] [CrossRef] [Green Version]
- Krumbhaar, E.B.; Krumbhaar, H.D. The Blood and Bone Marrow in Yelloe Cross Gas (Mustard Gas) Poisoning: Changes produced in the Bone Marrow of Fatal Cases. J. Med. Res. 1919, 40, 497–508. [Google Scholar] [PubMed]
- Goodman, L.S.; Wintrobe, M.; Dameshek, W.; Goodman, M.; Gilman, A.; McLennan, M. Landmark article Sept. 21, 1946: Nitrogen mustard therapy. Use of methyl-bis (beta-chloroethyl) amine hydrochloride and tris (beta-chloroethyl) amine hydrochloride for Hodgkin’s disease, lymphosarcoma, leukemia and certain allied and miscellaneous disorders. By Louis S. Goodman, Maxwell M. Wintrobe, William Dameshek, Morton J. Goodman, Alfred Gilman and Margaret T. McLennan. JAMA 1984, 251, 2255–2260. [Google Scholar] [CrossRef] [PubMed]
- Karnofsky, D.A.; Abelmann, W.H.; Craver, L.F.; Burchenal, J.H. The use of the nitrogen mustards in the palliative treatment of carcinoma. With particular reference to bronchogenic carcinoma. Cancer 1948, 1, 634–656. [Google Scholar] [CrossRef]
- Carter, S. Anticancer drug development progress: A comparison of approaches in the United States, the Soviet Union, Japan, and Western Europe. Natl. Cancer Inst. Monogr. 1974, 40, 31–42. [Google Scholar]
- Zaza, G.; Cheok, M.; Krynetskaia, N.; Thorn, C.; Stocco, G.; Hebert, J.M.; McLeod, H.; Weinshilboum, R.M.; Relling, M.V.; Evans, W.E.; et al. Thiopurine pathway. Pharmacogenetics Genom. 2010, 20, 573–574. [Google Scholar] [CrossRef]
- Parker, W.B. Enzymology of Purine and Pyrimidine Antimetabolites Used in the Treatment of Cancer. Chem. Rev. 2009, 109, 2880–2893. [Google Scholar] [CrossRef] [Green Version]
- Karran, P. Thiopurines, DNA damage, DNA repair and therapy-related cancer. Br. Med Bull. 2006, 79, 153–170. [Google Scholar] [CrossRef]
- Diouf, B.; Cheng, Q.; Krynetskaia, N.F.; Yang, W.; Cheok, M.; Pei, D.; Fan, Y.; Cheng, C.; Krynetskiy, E.Y.; Geng, H.; et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat. Med. 2011, 17, 1298–1303. [Google Scholar] [CrossRef]
- Fink, D.; Aebi, S.; Howell, S.B. The role of DNA mismatch repair in drug resistance. Clin. Cancer Res. 1998, 4, 1–6. [Google Scholar] [PubMed]
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-Fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, M.; Murray, M.S.; Kunkel, T.A.; Chou, K.-M. Interaction between DNA Polymerase λ and Anticancer Nucleoside Analogs. J. Biol. Chem. 2010, 285, 16874–16879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukushima, M.; Iizuka, K.; Jin, C.; Zhang, C.; Hong, M.; Eshima, K. Development of new promising antimetabolite, DFP-11207 with self-controlled toxicity in rodents. Drug Des. Dev. Ther. 2017, 11, 1693–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, W.-X.; Ditsworth, D.; Bauer, D.E.; Wang, Z.-Q.; Thompson, C.B. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004, 18, 1272–1282. [Google Scholar] [CrossRef] [Green Version]
- Peterson, D.E.; Sonis, S.T. Oral Complications of Cancer Chemotherapy; Springer Science and Business Media LLC: Cham, Switzerland, 1983; Volume 12. [Google Scholar]
- Fu, D.; Calvo, J.A.; Samson, L.D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat. Rev. Cancer 2012, 12, 104–120. [Google Scholar] [CrossRef] [Green Version]
- Kondo, N.; Takahashi, A.; Ono, K.; Ohnishi, T. DNA Damage Induced by Alkylating Agents and Repair Pathways. J. Nucleic Acids 2010, 2010, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, K.-I.; Sasaki, S.; Minoshima, M.; Bando, T.; Sugiyama, H. Alkylation of template strand of coding region causes effective gene silencing. Nucleic Acids Res. 2006, 34, 1189–1195. [Google Scholar] [CrossRef] [Green Version]
- Soll, J.M.; Sobol, R.W.; Mosammaparast, N. Regulation of DNA Alkylation Damage Repair: Lessons and Therapeutic Opportunities. Trends Biochem. Sci. 2017, 42, 206–218. [Google Scholar] [CrossRef] [Green Version]
- Dasari, S.; Tchounwou, P.B. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378. [Google Scholar] [CrossRef] [Green Version]
- Cameron, A.C.; Touyz, R.M.; Lang, N.N. Vascular Complications of Cancer Chemotherapy. Can. J. Cardiol. 2016, 32, 852–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nematbakhsh, M.; Pezeshki, Z.; Jazi, F.E.; Mazaheri, B.; Moeini, M.; Safari, T.; Azarkish, F.; Moslemi, F.; Maleki, M.; Rezaei, A.; et al. Cisplatin-Induced Nephrotoxicity; Protective Supplements and Gender Differences. Asian Pac. J. Cancer Prev. 2017, 18, 295–314. [Google Scholar] [PubMed]
- Bates, A.D.; Maxwell, A. Energy Coupling in Type II Topoisomerases: Why Do They Hydrolyze ATP? Biochem. 2007, 46, 7929–7941. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Sun, Y.; Ji, P.; Kopetz, S.; Zhang, W. Topoisomerase IIα in chromosome instability and personalized cancer therapy. Oncogene 2015, 34, 4019–4031. [Google Scholar] [CrossRef] [Green Version]
- Staker, B.L.; Hjerrild, K.; Feese, M.D.; Behnke, C.A.; Burgin, A.B.; Stewart, L. Nonlinear partial differential equations and applications: The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. USA 2002, 99, 15387–15392. [Google Scholar] [CrossRef] [Green Version]
- Pommier, Y. DNA Topoisomerase I Inhibitors: Chemistry, Biology, and Interfacial Inhibition. Chem. Rev. 2009, 109, 2894–2902. [Google Scholar] [CrossRef] [Green Version]
- McGowan, J.V.; Chung, R.; Maulik, A.; Piotrowska, I.; Walker, J.M.; Yellon, D.M. Anthracycline Chemotherapy and Cardiotoxicity. Cardiovasc. Drugs Ther. 2017, 31, 63–75. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.F.; Desai, S.D.; Li, T.-K.; Mao, Y.; Sun, M.; Sim, S.-P. Mechanism of Action of Camptothecin. Ann. N. Y. Acad. Sci. 2006, 922, 1–10. [Google Scholar] [CrossRef]
- Wu, J.; Liu, L.F. Processing of topoisomerase I cleavable complexes into DNA damage by transcription. Nucleic Acids Res. 1997, 25, 4181–4186. [Google Scholar] [CrossRef] [Green Version]
- Reyhanoglu, G.; Smith, T. Irinotecan. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Wallace, B.D.; Wang, H.; Lane, K.T.; Scott, J.E.; Orans, J.; Koo, J.S.; Venkatesh, M.; Jobin, C.; Yeh, L.-A.; Mani, S.; et al. Alleviating Cancer Drug Toxicity by Inhibiting a Bacterial Enzyme. Science 2010, 330, 831–835. [Google Scholar] [CrossRef] [Green Version]
- Chène, P.; Rudloff, J.; Schoepfer, J.; Furet, P.; Meier, P.; Qian, Z.; Schlaeppi, J.-M.; Schmitz, R.; Radimerski, T. Catalytic inhibition of topoisomerase II by a novel rationally designed ATP-competitive purine analogue. BMC Chem. Biol. 2009, 9, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Classen, S.; Olland, S.; Berger, J.M. Structure of the topoisomerase II ATPase region and its mechanism of inhibition by the chemotherapeutic agent ICRF-187. Proc. Natl. Acad. Sci. USA 2003, 100, 10629–10634, Erratum in Proc. Natl. Acad. Sci. USA 2003, 100, 14510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nitiss, J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer 2009, 9, 338–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wessel, I.; Jensen, L.H.; Jensen, P.B.; Falck, J.; Rose, A.; Roerth, M.; Nitiss, J.L.; Sehested, M. Human small cell lung cancer NYH cells selected for resistance to the bisdioxopiperazine topoisomerase II catalytic inhibitor ICRF-187 demonstrate a functional R162Q mutation in the Walker A consensus ATP binding domain of the α isoform. Cancer Res. 1999, 59, 3442–3450. [Google Scholar]
- Lee, J.H.; Wendorff, T.J.; Berger, J.M. Resveratrol: A novel type of topoisomerase II inhibitor. J. Biol. Chem. 2017, 292, 21011–21022. [Google Scholar] [CrossRef] [Green Version]
- Blatt, J. Second Malignancies Associated with Doxorubicin. Pediatr. Hematol. Oncol. 1995, 12, 111–113. [Google Scholar] [CrossRef]
- Brown, P.D.N.; for the Danish Society of Haematology Study Group on AML; Hoffmann, T.; Hansen, O.; Boesen, A.; Grønbæk, K.; Hippe, E.; Jensen, M.; Thorling, K.; Storm, H.; et al. Long-term survival and development of secondary malignancies in patients with acute myeloid leukemia treated with aclarubicin or daunorubicin plus cytosine arabinoside followed by intensive consolidation chemotherapy in a Danish national phase III trial. Leukemia 1997, 11, 37–41. [Google Scholar] [CrossRef]
- Kröger, N.; Damon, L.; Zander, A.R.; Wandt, H.; Derigs, G.; Ferrante, P.; Demirer, T.; Rosti, G.; Kr, N. Secondary acute leukemia following mitoxantrone-based high-dose chemotherapy for primary breast cancer patients. Bone Marrow Transplant. 2003, 32, 1153–1157. [Google Scholar] [CrossRef]
- Collins, I.; Weber, A.; Levens, D. Transcriptional Consequences of Topoisomerase Inhibition. Mol. Cell. Biol. 2001, 21, 8437–8451. [Google Scholar] [CrossRef] [Green Version]
- Cooper, G.M. Microtubules. In The Cell: A Molecular Approach; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Gigant, B.; Wang, C.; Ravelli, R.B.G.; Roussi, F.; Steinmetz, M.O.; Curmi, P.A.; Sobel, A.; Knossow, M. Structural basis for the regulation of tubulin by vinblastine. Nat. Cell Biol. 2005, 435, 519–522. [Google Scholar] [CrossRef]
- Jordan, M.A.; Himes, R.H.; Wilson, L. Comparison of the effects of vinblastine, vincristine, vindesine, and vinepidine on microtubule dynamics and cell proliferation in vitro. Cancer Res. 1985, 45, 2741–2747. [Google Scholar] [PubMed]
- Mhaidat, N.M.; Alzoubi, K.; Khabour, O.; Alawneh, K.; Raffee, L.; Alsatari, E.; Hussein, E.; Bani-Hani, K. Assessment of genotoxicity of vincristine, vinblastine and vinorelbine in human cultured lymphocytes: A comparative study. Balk. J. Med Genet. 2016, 19, 13–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, M.L.; Ojima, I. Chemistry and chemical biology of taxane anticancer agents. Chem. Rec. 2001, 1, 195–211. [Google Scholar] [CrossRef]
- Winefield, R.D.; Entwistle, R.A.; Foland, T.B.; Lushington, G.H.; Himes, R.H. Differences in Paclitaxel and Docetaxel Interactions with Tubulin Detected by Mutagenesis of Yeast Tubulin. ChemMedChem 2008, 3, 1844–1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verschraegen, C.F.; Sittisomwong, T.; Kudelka, A.P.; Guedes, E.D.P.; Steger, M.; Nelson-Taylor, T.; Vincent, M.; Rogers, R.; Atkinson, E.N.; Kavanagh, J.J. Docetaxel for Patients With Paclitaxel-Resistant Müllerian Carcinoma. J. Clin. Oncol. 2000, 18, 2733–2739. [Google Scholar] [CrossRef] [PubMed]
- Kenmotsu, H.; Tanigawara, Y. Pharmacokinetics, dynamics and toxicity of docetaxel: Why the Japanese dose differs from the Western dose. Cancer Sci. 2015, 106, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Azarenko, O.; Smiyun, G.; Mah, J.; Wilson, L.; Jordan, M.A. Antiproliferative Mechanism of Action of the Novel Taxane Cabazitaxel as Compared with the Parent Compound Docetaxel in MCF7 Breast Cancer Cells. Mol. Cancer Ther. 2014, 13, 2092–2103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vrignaud, P.; Sémiond, D.; Benning, V.; Beys, E.; Bouchard, H.; Gupta, S. Preclinical profile of cabazitaxel. Drug Des. Dev. Ther. 2014, 8, 1851–1867. [Google Scholar] [CrossRef]
- Pawlowska, E.; Szczepanska, J.; Szatkowska, M.; Blasiak, J. An Interplay between Senescence, Apoptosis and Autophagy in Glioblastoma Multiforme—Role in Pathogenesis and Therapeutic Perspective. Int. J. Mol. Sci. 2018, 19, 889. [Google Scholar] [CrossRef] [Green Version]
- Zeng, X.; Yan, T.; Schupp, J.E.; Seo, Y.; Kinsella, T.J. DNA Mismatch Repair Initiates 6-Thioguanine-Induced Autophagy through p53 Activation in Human Tumor Cells. Clin. Cancer Res. 2007, 13, 1315–1321. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Li, L.; Hu, J.; Zhao, Z.; Ji, L.; Cheng, C.; Zhang, G.; Zhang, T.; Li, Y.; Chen, H.; et al. UBL4A inhibits autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma via targeting LAMP1. J. Exp. Clin. Cancer Res. 2019, 38, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Vega-Rubín-De-Celis, S.; Zou, Z.; Fernández, Á.F.; Ci, B.; Kim, M.; Xiao, G.; Xie, Y.; Levine, B. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, 4176–4181. [Google Scholar]
- Kanzawa, T.; Germano, I.M.; Komata, T.; Ito, H.; Kondo, Y.; Kondo, S. Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ. 2004, 11, 448–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seto, T.; Sam, D.; Pan, M. Mechanisms of Primary and Secondary Resistance to Immune Checkpoint Inhibitors in Cancer. Med. Sci. 2019, 7, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Pascual, J.; Ayuso-Sacido, A.; Belda-Iniesta, C. Drug resistance in cancer immunotherapy: New strategies to improve checkpoint inhibitor therapies. Cancer Drug Resist. 2019, 2, 980–993. [Google Scholar] [CrossRef] [Green Version]
- Cui, Q.; Wang, J.-Q.; Assaraf, Y.G.; Ren, L.; Gupta, P.; Wei, L.; Ashby Jr, C.R.; Yang, D.-H.; Chen, Z.-S. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updat. 2018, 41, 1–25. [Google Scholar] [CrossRef]
- Kim, E.-K.; Jang, M.; Song, M.-J.; Kim, D.; Kim, Y.; Jang, H.H. Redox-Mediated Mechanism of Chemoresistance in Cancer Cells. Antioxidants 2019, 8, 471. [Google Scholar] [CrossRef] [Green Version]
- Machida, K. Existence of cancer stem cells in hepatocellular carcinoma: Myth or reality? Hepatol. Int. 2016, 11, 143–147. [Google Scholar] [CrossRef] [Green Version]
- Mindell, J.A. Lysosomal Acidification Mechanisms. Annu. Rev. Physiol. 2012, 74, 69–86. [Google Scholar] [CrossRef] [Green Version]
- Cooper, G.M. Lysosomes. In The Cell: A Molecular Approach; Sinauer Associates: Sunderland, MA, USA, 2000. [Google Scholar]
- Gorvel, J.-P.; Chavrier, P.; Zerial, M.; Gruenberg, J. rab5 controls early endosome fusion in vitro. Cell 1991, 64, 915–925. [Google Scholar] [CrossRef]
- Bucci, C.; Parton, R.G.; Mather, I.H.; Stunnenberg, H.; Simons, K.; Hoflack, B.; Zerial, M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 1992, 70, 715–728. [Google Scholar] [CrossRef] [Green Version]
- Christoforidis, S.; McBride, H.M.; Burgoyne, R.D.; Zerial, M. The Rab5 effector EEA1 is a core component of endosome docking. Nat. Cell Biol. 1999, 397, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Law, F.; Seo, J.H.; Wang, Z.; DeLeon, J.L.; Bolis, Y.; Brown, A.; Zong, W.-X.; Du, G.; Rocheleau, C.E. The VPS34 PI3K negatively regulates RAB-5 during endosome maturation. J. Cell Sci. 2017, 130, 2007–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 4th ed.; Transport from the trans Golgi network to lysosomes; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Thomas, D. Cell Biology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2017; Chapter 22—Endocytosis and the Endosomal Membrane System A2-Pollard; pp. 377–392. [Google Scholar]
- Lafourcade, C.; Sobo, K.; Kieffer-Jaquinod, S.; Garin, J.; Van Der Goot, G. Regulation of the V-ATPase along the Endocytic Pathway Occurs through Reversible Subunit Association and Membrane Localization. PLoS ONE 2008, 3, e2758. [Google Scholar] [CrossRef] [Green Version]
- Luzio, J.P.; Gray, S.R.; Bright, N.A. Endosome-lysosome fusion. Biochem. Soc. Trans. 2010, 38, 1413–1416. [Google Scholar] [CrossRef]
- Anandapadamanaban, M.; Masson, G.R.; Perisic, O.; Berndt, A.; Kaufman, J.; Johnson, C.M.; Santhanam, B.; Rogala, K.B.; Sabatini, D.M.; Williams, R.L. Architecture of human Rag GTPase heterodimers and their complex with mTORC1. Science 2019, 366, 203–210. [Google Scholar] [CrossRef]
- Martina, J.A.; Chen, Y.; Gucek, M.; Puertollano, R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 2012, 8, 903–914. [Google Scholar] [CrossRef] [Green Version]
- Medina, D.L.; Di Paola, S.; Peluso, I.; Armani, A.; De Stefani, D.; Venditti, R.; Montefusco, S.; Rosato, A.S.; Prezioso, C.; Forrester, A.; et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat. Cell Biol. 2015, 17, 288–299. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Dierichs, L.; Gu, J.-N.; Trajkovic-Arsic, M.; Hilger, R.A.; Savvatakis, K.; Vega-Rubin-De-Celis, S.; Liffers, S.-T.; Peña-Llopis, S.; Behrens, D.; et al. TFEB-mediated lysosomal biogenesis and lysosomal drug sequestration confer resistance to MEK inhibition in pancreatic cancer. Cell Death Discov. 2020, 6, 12–13. [Google Scholar] [CrossRef] [Green Version]
- Napolitano, G.; Di Malta, C.; Esposito, A.; de Araujo, M.E.; Pece, S.; Bertalot, G.; Matarese, M.; Benedetti, V.; Zampelli, A.; Stasyk, T. A substrate-specific mTORC1 pathway underlies Birt-Hogg-Dubé syndrome. Nature 2020, 585, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Perera, R.M.; Di Malta, C.; Ballabio, A. MiT/TFE Family of Transcription Factors, Lysosomes, and Cancer. Annu. Rev. Cancer Biol. 2019, 3, 203–222. [Google Scholar] [CrossRef]
- Yang, M.; Liu, E.; Tang, L.; Lei, Y.; Sun, X.; Hu, J.; Dong, H.; Yang, S.-M.; Gao, M.; Tang, B. Emerging roles and regulation of MiT/TFE transcriptional factors. Cell Commun. Signal. 2018, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ploper, D.; Taelman, V.F.; Robert, L.; Perez, B.S.; Titz, B.; Chen, H.-W.; Graeber, T.G.; Von Euw, E.; Ribas, A.; De Robertis, E. MITF drives endolysosomal biogenesis and potentiates Wnt signaling in melanoma cells. Proc. Natl. Acad. Sci. USA 2015, 112, E420–E429. [Google Scholar] [CrossRef] [Green Version]
- Martina, J.A.; Diab, H.I.; Lishu, L.; Jeong-A, L.; Patange, S.; Raben, N.; Puertollano, R. The Nutrient-Responsive Transcription Factor TFE3 Promotes Autophagy, Lysosomal Biogenesis, and Clearance of Cellular Debris. Sci. Signal. 2014, 7, ra9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annunziata, I.; Van De Vlekkert, D.; Wolf, E.; Finkelstein, D.; Neale, G.; Machado, E.; Mosca, R.; Campos, Y.; Tillman, H.; Roussel, M.F.; et al. MYC competes with MiT/TFE in regulating lysosomal biogenesis and autophagy through an epigenetic rheostat. Nat. Commun. 2019, 10, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Hämälistö, S.; Jäättelä, M. Lysosomes in cancer—Living on the edge (of the cell). Curr. Opin. Cell Biol. 2016, 39, 69–76. [Google Scholar] [CrossRef]
- Perera, R.M.; Stoykova, S.; Nicolay, B.N.; Ross, K.N.; Fitamant, J.; Boukhali, M.; Lengrand, J.; Deshpande, V.; Selig, M.K.; Ferrone, C.R.; et al. Transcriptional control of autophagy–lysosome function drives pancreatic cancer metabolism. Nat. Cell Biol. 2015, 524, 361–365. [Google Scholar] [CrossRef]
- Rodrigo, M.A.M.; Buchtelova, H.; Rios, V.D.L.; Casal, J.I.; Eckschlager, T.; Hrabeta, J.; Belhajova, M.; Hegerab, Z.; Adamab, V. Proteomic Signature of Neuroblastoma Cells UKF-NB-4 Reveals Key Role of Lysosomal Sequestration and the Proteasome Complex in Acquiring Chemoresistance to Cisplatin. J. Proteome Res. 2018, 18, 1255–1263. [Google Scholar] [CrossRef] [Green Version]
- Levy, J.M.M.; Zahedi, S.; Griesinger, A.M.; Morin, A.; Davies, K.D.; Aisner, D.L.; Kleinschmidt-DeMasters, B.K.; E Fitzwalter, B.; Goodall, M.L.; Thorburn, J.; et al. Autophagy inhibition overcomes multiple mechanisms of resistance to BRAF inhibition in brain tumors. eLife 2017, 6, e19671. [Google Scholar] [CrossRef]
- Zhitomirsky, B.; Yunaev, A.; Kreiserman, R.; Kaplan, A.; Stark, M.; Assaraf, Y.G. Lysosomotropic drugs activate TFEB via lysosomal membrane fluidization and consequent inhibition of mTORC1 activity. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Tam, A.; Santi, S.A.; Parissenti, A.M. Role of autophagy and lysosomal drug sequestration in acquired resistance to doxorubicin in MCF-7 cells. BMC Cancer 2016, 16, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolgova, N.V.; Yu, C.; Cvitkovic, J.P.; Hodak, M.; Nienaber, K.H.; Summers, K.L.; Cotelesage, J.J.H.; Bernholc, J.; Kaminski, G.A.; Pickering, I.J.; et al. Binding of Copper and Cisplatin to Atox1 Is Mediated by Glutathione through the Formation of Metal–Sulfur Clusters. Biochemistry 2017, 56, 3129–3141. [Google Scholar] [CrossRef] [PubMed]
- Petruzzelli, R.; Polishchuk, R. Activity and Trafficking of Copper-Transporting ATPases in Tumor Development and Defense against Platinum-Based Drugs. Cells 2019, 8, 1080. [Google Scholar] [CrossRef] [Green Version]
- Polishchuk, R.; Polishchuk, R.S. The emerging role of lysosomes in copper homeostasis. Metallomics 2016, 8, 853–862. [Google Scholar] [CrossRef]
- Lukanović, D.; Herzog, M.; Kobal, B.; Černe, K. The contribution of copper efflux transporters ATP7A and ATP7B to chemoresistance and personalized medicine in ovarian cancer. Biomed. Pharmacother. 2020, 129, 110401. [Google Scholar] [CrossRef]
- Kalayda, G.V.; Wagner, C.H.; Buss, I.; Reedijk, J.; Jaehde, U. Altered localisation of the copper efflux transporters ATP7A and ATP7B associated with cisplatin resistance in human ovarian carcinoma cells. BMC Cancer 2008, 8, 175. [Google Scholar] [CrossRef] [Green Version]
- Aits, S.; Jäättelä, M. Lysosomal cell death at a glance. J. Cell Sci. 2013, 126, 1905–1912. [Google Scholar] [CrossRef] [Green Version]
- Sukhai, M.A.; Prabha, S.; Hurren, R.; Rutledge, A.C.; Lee, A.Y.; Sriskanthadevan, S.; Sun, H.; Wang, X.; Skrtic, M.; Seneviratne, A.; et al. Lysosomal disruption preferentially targets acute myeloid leukemia cells and progenitors. J. Clin. Investig. 2013, 123, 315–328. [Google Scholar] [CrossRef]
- Wang, Q.; Yao, J.; Jin, Q.; Wang, X.; Zhu, H.; Huang, F.; Wang, W.; Qiang, J.; Ni, Q. LAMP1 expression is associated with poor prognosis in breast cancer. Oncol. Lett. 2017, 14, 4729–4735. [Google Scholar] [CrossRef] [Green Version]
- Sarafian, V.S.; Koev, I.G.; Mehterov, N.; Kazakova, M.; Dangalov, K. LAMP-1 gene is overexpressed in high grade glioma. APMIS 2018, 126, 657–662. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.; van de Vlekkert, D.; Annunziata, I.; d’Azzo, A. Regulated lysosomal exocytosis mediates cancer progression. Mol. Genet. Metab. 2016, 117, S76. [Google Scholar] [CrossRef] [Green Version]
- Damaghi, M.; Tafreshi, N.K.; Lloyd, M.C.; Sprung, R.; Estrella, V.; Wojtkowiak, J.W.; Morse, D.L.; Koomen, J.M.; Bui, M.M.; A Gatenby, R.; et al. Chronic acidosis in the tumour microenvironment selects for overexpression of LAMP2 in the plasma membrane. Nat. Commun. 2015, 6, 8752. [Google Scholar] [CrossRef] [Green Version]
- Sennoune, S.R.; Bakunts, K.; Martínez, G.M.; Chua-Tuan, J.L.; Kebir, Y.; Attaya, M.N.; Martínez-Zaguilán, R. Vacuolar H+-ATPase in human breast cancer cells with distinct metastatic potential: Distribution and functional activity. Am. J. Physiol. Physiol. 2004, 286, C1443–C1452. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.W. Solving the secretory acid sphingomyelinase puzzle: Insights from lysosome--mediated parasite invasion and plasma membrane repair. Cell. Microbiol. 2019, 21, e13065. [Google Scholar] [CrossRef] [PubMed]
- Rebillard, A.; Rioux-Leclercq, N.; Müller, C.; Bellaud, P.; Jouan, F.; Meurette, O.; Jouan, E.; Vernhet, L.; Le Quément, C.; Carpinteiro, A.; et al. Acid sphingomyelinase deficiency protects from cisplatin-induced gastrointestinal damage. Oncogene 2008, 27, 6590–6595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Jiang, Q.; Zhu, P.; Chen, Y.; Xie, X.; Du, Z.; Jiang, L.; Tang, W. NPRL2 enhances autophagy and the resistance to Everolimus in castration-resistant prostate cancer. Prostate 2019, 79, 44–53. [Google Scholar] [CrossRef] [Green Version]
- Harwood, F.C.; Geltink, R.I.K.; O’Hara, B.P.; Cardone, M.; Janke, L.; Finkelstein, D.; Entin, I.; Paul, L.; Houghton, P.J.; Grosveld, G. ETV7 is an essential component of a rapamycin-insensitive mTOR complex in cancer. Sci. Adv. 2018, 4, eaar3938. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, A.M.; Krise, J.P. Lysosomal Sequestration of Amine-Containing Drugs: Analysis and Therapeutic Implications. J. Pharm. Sci. 2007, 96, 729–746. [Google Scholar] [CrossRef]
- Derendorf, H. Excessive lysosomal ion-trapping of hydroxychloroquine and azithromycin. Int. J. Antimicrob. Agents 2020, 55, 106007. [Google Scholar] [CrossRef]
- Stark, M.; Silva, T.F.D.; Levin, G.; Machuqueiro, M.; Assaraf, Y.G. The Lysosomotropic Activity of Hydrophobic Weak Base Drugs is Mediated via Their Intercalation into the Lysosomal Membrane. Cells 2020, 9, 1082. [Google Scholar] [CrossRef] [PubMed]
- Boya, P.; Kroemer, G. Lysosomal membrane permeabilization in cell death. Oncogene 2008, 27, 6434–6451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dielschneider, R.F.; Henson, E.S.; Gibson, S.B. Lysosomes as Oxidative Targets for Cancer Therapy. Oxidative Med. Cell. Longev. 2017, 2017, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhitomirsky, B.; Assaraf, Y.G. Lysosomal accumulation of anticancer drugs triggers lysosomal exocytosis. Oncotarget 2017, 8, 45117–45132. [Google Scholar] [CrossRef] [Green Version]
- Von Schwarzenberg, K.; Lajtos, T.; Simon, L.; Müller, R.; Vereb, G.; Vollmar, A.M. V-ATPase inhibition overcomes trastuzumab resistance in breast cancer. Mol. Oncol. 2014, 8, 9–19. [Google Scholar] [CrossRef]
- Whitton, B.; Okamoto, H.; Packham, G.; Crabb, S.J. Vacuolar ATPase as a potential therapeutic target and mediator of treatment resistance in cancer. Cancer Med. 2018, 7, 3800–3811. [Google Scholar] [CrossRef]
- Goldman, S.D.B.; Funk, R.S.; A Rajewski, R.; Krise, J.P. Mechanisms of amine accumulation in, and egress from, lysosomes. Bioanalysis 2009, 1, 1445–1459. [Google Scholar] [CrossRef] [Green Version]
- Henics, T.; Wheatley, D.N. Cytoplasmic vacuolation, adaptation and cell death: A view on new perspectives and features. Biol. Cell 1999, 91, 485–498. [Google Scholar] [CrossRef]
- Zhang, X.; Cheng, X.; Yu, L.; Yang, J.; Calvo, R.; Patnaik, S.; Hu, X.; Gao, Q.; Yang, M.; Lawas, M.; et al. MCOLN1 is a ROS sensor in lysosomes that regulates autophagy. Nat. Commun. 2016, 7, 12109. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, A.M.; Krise, J.P. Niemann-Pick C1 Functions in Regulating Lysosomal Amine Content. J. Biol. Chem. 2008, 283, 24584–24593. [Google Scholar] [CrossRef] [Green Version]
- Zhitomirsky, B.; Assaraf, Y.G. Lysosomes as mediators of drug resistance in cancer. Drug Resist. Updat. 2016, 24, 23–33. [Google Scholar] [CrossRef] [PubMed]
- De Klerk, D.; Honeywell, R.J.; Jansen, G.; Peters, G.J. Transporter and Lysosomal Mediated (Multi)drug Resistance to Tyrosine Kinase Inhibitors and Potential Strategies to Overcome Resistance. Cancers 2018, 10, 503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, F.S.; Trombetta, E.; Cetrangolo, P.; Maggioni, M.; Razini, P.; De Santis, F.; Torrente, Y.; Prati, D.; Torresani, E.; Porretti, L. Giant Lysosomes as a Chemotherapy Resistance Mechanism in Hepatocellular Carcinoma Cells. PLoS ONE 2014, 9, e114787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamagishi, T.; Sahni, S.; Sharp, D.M.; Arvind, A.; Jansson, P.J.; Richardson, D.R. P-glycoprotein Mediates Drug Resistance via a Novel Mechanism Involving Lysosomal Sequestration. J. Biol. Chem. 2013, 288, 31761–31771. [Google Scholar] [CrossRef] [Green Version]
- Lu, S.; Sung, T.; Lin, N.; Abraham, R.T.; Jessen, B.A. Lysosomal adaptation: How cells respond to lysosomotropic compounds. PLoS ONE 2017, 12, e0173771. [Google Scholar] [CrossRef] [Green Version]
- Zhitomirsky, B.; Assaraf, Y.G. Lysosomal sequestration of hydrophobic weak base chemotherapeutics triggers lysosomal biogenesis and lysosome-dependent cancer multidrug resistance. Oncotarget 2014, 6, 1143–1156. [Google Scholar] [CrossRef] [Green Version]
- Vincent, L.-A.; Attaoua, C.; Bellis, M.; Rozkydalova, L.; Vian, L.; Cuq, P.; Hadj-Kaddour, K. Lysosomes and unfolded protein response, determinants of differential resistance of melanoma cells to vinca alkaloids. Fundam. Clin. Pharmacol. 2015, 29, 164–177. [Google Scholar] [CrossRef]
- A Seebacher, N.; Richardson, D.R.; Jansson, P.J. A mechanism for overcoming P-glycoprotein-mediated drug resistance: Novel combination therapy that releases stored doxorubicin from lysosomes via lysosomal permeabilization using Dp44mT or DpC. Cell Death Dis. 2016, 7, e2510. [Google Scholar] [CrossRef]
- Shao, M.; Zhu, W.; Lv, X.; Yang, Q.; Liu, X.; Xie, Y.; Tang, P.; Sun, L. Encapsulation of chloroquine and doxorubicin by MPEG-PLA to enhance anticancer effects by lysosomes inhibition in ovarian cancer. Int. J. Nanomed. 2018, 13, 8231–8245. [Google Scholar] [CrossRef] [Green Version]
- Salaroglio, I.C.; Gazzano, E.; Abdullrahman, A.; Mungo, E.; Castella, B.; Abd-Elrahman, G.E.; Massaia, M.; Donadelli, M.; Rubinstein, M.; Riganti, C.; et al. Increasing intratumor C/EBP-β LIP and nitric oxide levels overcome resistance to doxorubicin in triple negative breast cancer. J. Exp. Clin. Cancer Res. 2018, 37, 286. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Y.; Jing, L.; Wang, J.; Yan, Y.; Feng, Y.; Zhang, Y.; Liu, Z.; Ma, L.; Diao, A. Lysosome Inhibitors Enhance the Chemotherapeutic Activity of Doxorubicin in HepG2 Cells. Chemotherapy 2016, 62, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Agostinis, P.; Berg, K.; Cengel, K.A.; Foster, T.H.; Girotti, A.W.; Gollnick, S.O.; Hahn, S.M.; Hamblin, M.R.; Juzeniene, A.; Kessel, D.; et al. Photodynamic therapy of cancer: An update. CA Cancer J. Clin. 2011, 61, 250–281. [Google Scholar] [CrossRef] [PubMed]
- Domagala, A.; Fidyt, K.; Bobrowicz, M.; Stachura, J.; Szczygiel, K.; Firczuk, M. Typical and Atypical Inducers of Lysosomal Cell Death: A Promising Anticancer Strategy. Int. J. Mol. Sci. 2018, 19, 2256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Granato, M.; Lacconi, V.; Peddis, M.; Lotti, L.V.; Di Renzo, L.M.; Gonnella, R.; Santarelli, R.L.; Trivedi, P.; Frati, L.; Orazi, G.D.; et al. HSP70 inhibition by 2-phenylethynesulfonamide induces lysosomal cathepsin D release and immunogenic cell death in primary effusion lymphoma. Cell Death Dis. 2013, 4, e730. [Google Scholar] [CrossRef] [PubMed]
- Monma, H.; Harashima, N.; Inao, T.; Okano, S.; Tajima, Y.; Harada, M. The HSP70 and autophagy inhibitor pifithrin-μ enhances the antitumor effects of TRAIL on human pancreatic cancer. Mol. Cancer Ther. 2013, 12, 341–351. [Google Scholar] [CrossRef] [Green Version]
- Mai, T.T.; Hamaï, A.; Hienzsch, A.; Cañeque, T.; Müller, S.; Wicinski, J.; Cabaud, O.; Leroy, C.; David, A.; Acevedo, V.; et al. Salinomycin kills cancer stem cells by sequestering iron in lysosomes. Nat. Chem. 2017, 9, 1025–1033. [Google Scholar] [CrossRef] [Green Version]
- Ridinger, J.; Koeneke, E.; Kolbinger, F.R.; Koerholz, K.; Mahboobi, S.; Hellweg, L.; Gunkel, N.; Miller, A.K.; Peterziel, H.; Schmezer, P.; et al. Dual role of HDAC10 in lysosomal exocytosis and DNA repair promotes neuroblastoma chemoresistance. Sci. Rep. 2018, 8, 10039. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Kumar, S.; Prasad, D.; Bhardwaj, T. Therapeutic journery of nitrogen mustard as alkylating anticancer agents: Historic to future perspectives. Eur. J. Med. Chem. 2018, 151, 401–433. [Google Scholar] [CrossRef]
- Uboha, N.; Hochster, H.S. TAS-102: A novel antimetabolite for the 21st century. Future Oncol. 2016, 12, 153–163. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Dhillon, S.; Deeks, E.D. Trifluridine/Tipiracil: A Review in Metastatic Gastric Cancer. Drugs 2019, 79, 1583–1590. [Google Scholar] [CrossRef] [Green Version]
- Chattopadhyay, S.; Moran, R.G.; Goldman, I.D. Pemetrexed: Biochemical and cellular pharmacology, mechanisms, and clinical applications. Mol. Cancer Ther. 2007, 6, 404–417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joerger, M. Cancer chemotherapy: Targeting folic acid synthesis. Cancer Manag. Res. 2010, 2, 293. [Google Scholar] [CrossRef] [Green Version]
- Khan, Q.A.; Pilch, D.S. Topoisomerase I-mediated DNA Cleavage Induced by the Minor Groove-directed Binding of Bibenzimidazoles to a Distal Site. J. Mol. Biol. 2007, 365, 561–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parness, J.; Horwitz, S.B. Taxol binds to polymerized tubulin in vitro. J. Cell Biol. 1981, 91, 479–487. [Google Scholar] [CrossRef] [PubMed]

| Drug Family | Drug Class | Drug Name | Molecular Mode of Action | References |
|---|---|---|---|---|
| Alkylating Agents | Nitrogen mustard | N-methyl-bis(2-chloroethyl) amines, Chlorambucil, Melphalan, Cyclophosphamide | Crosslinks DNA strands through initiating nucleophilic substitution with reactive centers on N and O of DNA base substrates | [15,134] |
| Temozolomide, N-methyl-N-Nitrosoguanidine, Procarbazine | Forms localized N and O-alkyl adduct on one reactive center of DNA substrates that serves as the basis for extensive genotoxic damages, including DNA double-stranded breaks and chromosomal translocation. | [17,18,19] | ||
| Platinum-based alkylating agent | Cisplatin | Induces DNA intra-strand crosslinks, reactive oxygen species production, cell cycle arrest, and alterations of Ca2+ signaling | [21] | |
| Antimetabolites | Purine antagonist | Mercaptopurine (6-MP) | Inhibits de novo purine synthesis Produces genotoxic thioguanine nucleotides that inhibit DNA and RNA synthesis and alter their subsequent metabolism | [7,9] |
| Thymidine phophorylase inhibitor | Tipiracil Hydrochloride (TAS-102) | Induces cell cycle arrest at G2 phase and DNA double-strand breaks with enhanced drug potency. | [135,136] | |
| Antifolate | Methotrexate, Pemetrexed | Depletes intracellular thymidine by inhibiting TYMS, DHFR, AICARFT, GART Increases intracellular dUMP and promotes its misincorporation into the DNA in inducing double-strand breaks. | [137,138] | |
| Topoisomerase Inhibitors | Topo I inhibitor | Camptothecin | Uncompetitively cleaves DNA-topo 1 binary complex and enlarges the DNA cleavage gap, preventing further DNA re-ligation while causing collision with replication forks and eliciting apoptosis through exposure of DNA free ends. | [26,27] |
| Bibenzimidazole, Terbenzimidazole | Poisons Topo I by binding to the DNA minor groove | [139] | ||
| Topo II inhibitor | Doxorubicin, Daunorubicin, Idarubicin | Intercalates DNA and forms DNA covalent adducts Mediates Cell cycle arrest Generates reactive oxidative species | [28] | |
| Bisdioxopiperazine | Stabilizes ATP-bound Topo II, thereby inhibiting the enzyme’s catalytic cycles | [34,35] | ||
| Resveratrol | Prevents dimerization of Topo II ATPase domain, thereby allosterically inhibiting the enzyme’s catalytic cycles | [37] | ||
| Microtubule-targeting Agents | Vinca alkaloid | Vincristine, Vinblastine, Vinorelbine | Inhibits microtubule polymerization Induces oxidative DNA damage | [43,45] |
| Taxane | Docetaxel, Paclitaxel | Binds to and stabilizes tubulin polymers | [140] | |
| Cabazitaxel | Inhibits microtubule shortening and overall dynamicity | [50,51] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, X.; El Hiani, Y. Getting Lost in the Cell–Lysosomal Entrapment of Chemotherapeutics. Cancers 2020, 12, 3669. https://doi.org/10.3390/cancers12123669
Zhai X, El Hiani Y. Getting Lost in the Cell–Lysosomal Entrapment of Chemotherapeutics. Cancers. 2020; 12(12):3669. https://doi.org/10.3390/cancers12123669
Chicago/Turabian StyleZhai, Xingjian, and Yassine El Hiani. 2020. "Getting Lost in the Cell–Lysosomal Entrapment of Chemotherapeutics" Cancers 12, no. 12: 3669. https://doi.org/10.3390/cancers12123669