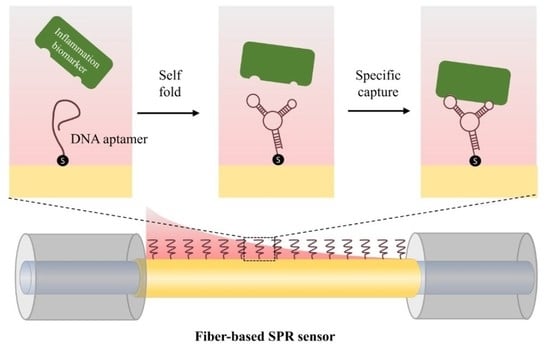
A Fiber-Based SPR Aptasensor for the In Vitro Detection of Inflammation Biomarkers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
- −
- Binding buffer: 20 mM tris base, 1 mM EDTA, 1 mM TCEP, pH = 8.0
- −
- Detection buffer: 25 mM tris base, 192 mM glycine, 0.05% Tween 20, 0.1 w/t% BSA, 2 mM CaCl2, 100 mM NaCl, pH = 8.0
- −
- Washing buffer: 20 mM tris base, 1 mM EDTA, 0.001% SDS, pH = 8.0
2.2. Fiber-Based Biosensor Fabrication and System Set-Up
3. Results and Discussion
3.1. The Characterization of the Sensing Areas
3.2. Basic Performance Testing of the SPR Biosensor
3.3. CRP and cTn-I Detection
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beaglehole, R. International Trends in Coronary Heart Disease Mortality, Morbidity, and Risk Factors. Epidemiol. Rev. 1990, 12, 1–15. [Google Scholar] [CrossRef]
- Chang, W.H.; Mueller, S.H.; Chung, S.-C.; Foster, G.R.; Lai, A.G. Increased Burden of Cardiovascular Disease in People with Liver Disease: Unequal Geographical Variations, Risk Factors and Excess Years of Life Lost. J. Transl. Med. 2022, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, S.; Huang, A.; Ono, Y.; Rajan, S. Continuous Artery Monitoring Using a Flexible and Wearable Single-Element Ultrasonic Sensor. IEEE Instrum. Meas. Mag. 2022, 25, 6–11. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Hoshino, M.; Sugiyama, T.; Kanaji, Y.; Nagamine, T.; Misawa, T.; Hada, M.; Araki, M.; Hamaya, R.; Usui, E.; et al. Association of Near-Infrared Spectroscopy-Defined Lipid Rich Plaque with Lesion Morphology and Peri-Coronary Inflammation on Computed Tomography Angiography. Atherosclerosis 2022, 346, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Anzai, T.; Kaneko, H.; Mano, Y.; Anzai, A.; Maekawa, Y.; Takahashi, T.; Meguro, T.; Yoshikawa, T.; Fukuda, K. C-Reactive Protein Overexpression Exacerbates Pressure Overload–Induced Cardiac Remodeling Through Enhanced Inflammatory Response. Hypertension 2011, 57, 208–215. [Google Scholar] [CrossRef]
- Haverkate, E.; Thompson, S.G.; Pyke, S.D.; Gallimore, J.R.; Group, M.B.P. Production of C-Reactive Protein and Risk of Coronary Events in Stable and Unstable Angina. Lancet 1997, 349, 462–466. [Google Scholar] [CrossRef]
- Saenger, A.K.; Korpi-Steiner, N. Chapter One—Advances in Cardiac Biomarkers of Acute Coronary Syndrome. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Academic Press: New York, NY, USA, 2017; Volume 78, pp. 1–58. ISBN 978-0-12-811919-8. [Google Scholar]
- Bozkurt, B. Chapter 24—Heart Failure as a Consequence of Dilated Cardiomyopathy. In Heart Failure: A Companion to Braunwald’s Heart Disease, 2nd ed.; Mann, D.L., Ed.; W.B. Saunders: Philadelphia, PA, USA, 2011; pp. 372–394. ISBN 978-1-4160-5895-3. [Google Scholar]
- Rathnayake, N.; Åkerman, S.; Klinge, B.; Lundegren, N.; Jansson, H.; Tryselius, Y.; Sorsa, T.; Gustafsson, A. Salivary Biomarkers for Detection of Systemic Diseases. PLoS ONE 2013, 8, e61356. [Google Scholar] [CrossRef] [Green Version]
- Pinyorospathum, C.; Chaiyo, S.; Sae-ung, P.; Hoven, V.P.; Damsongsang, P.; Siangproh, W.; Chailapakul, O. Disposable Paper-Based Electrochemical Sensor Using Thiol-Terminated Poly (2-Methacryloyloxyethyl Phosphorylcholine) for the Label-Free Detection of C-Reactive Protein. Microchim. Acta 2019, 186, 472. [Google Scholar] [CrossRef]
- Naito, Y.; Takagi, T.; Okada, H.; Omatsu, T.; Mizushima, K.; Handa, O.; Kokura, S.; Ichikawa, H.; Fujiwake, H.; Yoshikawa, T. Identification of Inflammation-Related Proteins in a Murine Colitis Model by 2D Fluorescence Difference Gel Electrophoresis and Mass Spectrometry. J. Gastroenterol. Hepatol. 2010, 25, 144–148. [Google Scholar] [CrossRef]
- Wignarajah, S.; Suaifan, G.A.R.Y.; Bizzarro, S.; Bikker, F.J.; Kaman, W.E.; Zourob, M. Colorimetric Assay for the Detection of Typical Biomarkers for Periodontitis Using a Magnetic Nanoparticle Biosensor. Anal. Chem. 2015, 87, 12161–12168. [Google Scholar] [CrossRef]
- Myszka, D.G. Kinetic, Equilibrium, and Thermodynamic Analysis of Macromolecular Interactions with BIACORE. Methods Enzymol. 2000, 323, 325–340. [Google Scholar] [CrossRef] [PubMed]
- Winzor, D.J. Interpretation of Results from the Competitive Biacore Procedure for Characterizing Immunochemical Interactions in Solution. J. Mol. Recognit. 2018, 31, 270–274. [Google Scholar] [CrossRef] [PubMed]
- Hoa, X.D.; Kirk, A.G.; Tabrizian, M. Towards Integrated and Sensitive Surface Plasmon Resonance Biosensors: A Review of Recent Progress. Biosens. Bioelectron. 2007, 23, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Prabowo, B.A.; Chang, Y.-F.; Lee, Y.-Y.; Su, L.-C.; Yu, C.-J.; Lin, Y.-H.; Chou, C.; Chiu, N.-F.; Lai, H.-C.; Liu, K.-C. Application of an OLED Integrated with BEF and Giant Birefringent Optical (GBO) Film in a SPR Biosensor. Sens. Actuators B Chem. 2014, 198, 424–430. [Google Scholar] [CrossRef]
- Masson, J.-F.; Battaglia, T.M.; Beaudoin, S.; Booksh, K.S. Applications of Small Surface Plasmon Resonance Sensors for Biochemical Monitoring. In Proceedings of the Smart Medical and Biomedical Sensor Technology II, SPIE, Philadelphia, PA, USA, 17 December 2004; Volume 5588, pp. 178–191. [Google Scholar]
- Horta, S.; Qu, J.-H.; Dekimpe, C.; Bonnez, Q.; Vandenbulcke, A.; Tellier, E.; Kaplanski, G.; Delport, F.; Geukens, N.; Lammertyn, J.; et al. Co(III)-NTA Mediated Antigen Immobilization on a Fiber Optic-SPR Biosensor for Detection of Autoantibodies in Autoimmune Diseases: Application in Immune-Mediated Thrombotic Thrombocytopenic Purpura. Anal. Chem. 2020, 92, 13880–13887. [Google Scholar] [CrossRef]
- Stamatis, C.; Farid, S.S. Process Economics Evaluation of Cell-Free Synthesis for the Commercial Manufacture of Antibody Drug Conjugates. Biotechnol. J. 2021, 16, 2000238. [Google Scholar] [CrossRef]
- Pabari, R.M.; Ryan, B.; McCarthy, C.; Ramtoola, Z. Effect of Microencapsulation Shear Stress on the Structural Integrity and Biological Activity of a Model Monoclonal Antibody, Trastuzumab. Pharmaceutics 2011, 3, 510–524. [Google Scholar] [CrossRef] [Green Version]
- Bostrom, J.; Lee, C.V.; Haber, L.; Fuh, G. Improving Antibody Binding Affinity and Specificity for Therapeutic Development. In Therapeutic Antibodies: Methods and Protocols; Methods in Molecular BiologyTM; Dimitrov, A.S., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 525, pp. 353–376. ISBN 978-1-59745-554-1. [Google Scholar]
- Zhu, Q.; Liu, G.; Kai, M. DNA Aptamers in the Diagnosis and Treatment of Human Diseases. Molecules 2015, 20, 20979–20997. [Google Scholar] [CrossRef]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2020, 25, 3. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Jiang, R.; Wang, Q.; Huang, J.; Yang, X.; Wang, K.; Li, W.; Chen, N.; Li, Q. Detection of C-Reactive Protein Using Nanoparticle-Enhanced Surface Plasmon Resonance Using an Aptamer-Antibody Sandwich Assay. Chem. Commun. 2016, 52, 3568–3571. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Lin, X.; Lu, J.; Wei, P.; Luo, Z.; Lu, X.; Chen, Z.; Zhang, L. DNA Nanotetrahedron-Assisted Electrochemical Aptasensor for Cardiac Troponin I Detection Based on the Co-Catalysis of Hybrid Nanozyme, Natural Enzyme and Artificial DNAzyme. Biosens. Bioelectron. 2019, 142, 111578. [Google Scholar] [CrossRef]
- Lu, B.; Sun, Y.; Lai, X.; Pu, Z.; Yu, H.; Xu, K.; Li, D. Side-Polished Fiber SPR Sensor with Tempetrature Self-Compensation for Continuous Glucose Monitoring. In Proceedings of the 2016 IEEE 29th International Conference on Micro Electro Mechanical Systems (MEMS), Shanghai, China, 24 January 2016; pp. 411–414. [Google Scholar]
- Lu, B.; Lai, X.; Zhang, P.; Wu, H.; Yu, H.; Li, D. Roughened Cylindrical Gold Layer with Curve Graphene Coating for Enhanced Sensitivity of Fiber SPR Sensor. In Proceedings of the 2017 19th International Conference on Solid-State Sensors, Actuators and Microsystems (TRANSDUCERS), Kaohsiung, Taiwan, 18 June 2017; pp. 1991–1994. [Google Scholar]
- Hoyt, L.F. New Table of the Refractive Index of Pure Glycerol at 20 °C. Ind. Eng. Chem. 1934, 26, 329–332. [Google Scholar] [CrossRef]
- Walter, J.-G.; Kökpinar, Ö.; Friehs, K.; Stahl, F.; Scheper, T. Systematic Investigation of Optimal Aptamer Immobilization for Protein−Microarray Applications. Anal. Chem. 2008, 80, 7372–7378. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhao, L.; Wang, Y.; Hameed, S.; Ping, J.; Xie, L.; Ying, Y. Recent Advances in Food-Derived Nanomaterials Applied to Biosensing. TrAC Trends Anal. Chem. 2020, 127, 115884. [Google Scholar] [CrossRef]
- Gao, S.; Koshizaki, N.; Tokuhisa, H.; Koyama, E.; Sasaki, T.; Kim, J.-K.; Ryu, J.; Kim, D.-S.; Shimizu, Y. Highly Stable Au Nanoparticles with Tunable Spacing and Their Potential Application in Surface Plasmon Resonance Biosensors. Adv. Funct. Mater. 2010, 20, 78–86. [Google Scholar] [CrossRef]
- Qu, L.; Xu, J.; Tan, X.; Liu, Z.; Xu, L.; Peng, R. Dual-Aptamer Modification Generates a Unique Interface for Highly Sensitive and Specific Electrochemical Detection of Tumor Cells. ACS Appl. Mater. Interfaces 2014, 6, 7309–7315. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Liu, J.; Zhang, P.; Li, J.; Ji, H.; Yang, X.; Wang, K. A Recognition-before-Labeling Strategy for Sensitive Detection of Lung Cancer Cells with a Quantum Dot–Aptamer Complex. Analyst 2015, 140, 6100–6107. [Google Scholar] [CrossRef]
- Yang, X.-H.; Kong, W.-J.; Yang, M.-H.; Zhao, M.; Ouyang, Z. Application of Aptamer Identification Technology in Rapid Analysis of Mycotoxins. Chin. J. Anal. Chem. 2013, 41, 297–306. [Google Scholar] [CrossRef]
- Oberthür, D.; Achenbach, J.; Gabdulkhakov, A.; Buchner, K.; Maasch, C.; Falke, S.; Rehders, D.; Klussmann, S.; Betzel, C. Crystal Structure of a Mirror-Image L-RNA Aptamer (Spiegelmer) in Complex with the Natural L-Protein Target CCL2. Nat. Commun. 2015, 6, 6923. [Google Scholar] [CrossRef] [Green Version]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for Improving Aptamer Binding Affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef]
- Zhao, W.; Chiuman, W.; Lam, J.C.F.; McManus, S.A.; Chen, W.; Cui, Y.; Pelton, R.; Brook, M.A.; Li, Y. DNA Aptamer Folding on Gold Nanoparticles: From Colloid Chemistry to Biosensors. J. Am. Chem. Soc. 2008, 130, 3610–3618. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Phares, N.; Lubin, A.A.; Xiao, Y.; Plaxco, K.W. Optimization of Electrochemical Aptamer-Based Sensors via Optimization of Probe Packing Density and Surface Chemistry. Langmuir 2008, 24, 10513–10518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frederix, F.; Bonroy, K.; Laureyn, W.; Reekmans, G.; Campitelli, A.; Dehaen, W.; Maes, G. Enhanced Performance of an Affinity Biosensor Interface Based on Mixed Self-Assembled Monolayers of Thiols on Gold. Langmuir 2003, 19, 4351–4357. [Google Scholar] [CrossRef]






| Aptamer Type | The Sequence (5′ to 3′) |
|---|---|
| CRP | GGCAGGAAGACAAACACGATGGGGGGGTATGATTTGATGTGGTTGTGCATGATCGTGGTCTGTGGTGCTGTTTTT |
| cTn-I | CGAAGGGGATTCGAGGGGTGATTGCGTGCTCCATTTGGTGTTTTT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Y.; Wang, R.; Li, D. A Fiber-Based SPR Aptasensor for the In Vitro Detection of Inflammation Biomarkers. Micromachines 2022, 13, 1036. https://doi.org/10.3390/mi13071036
Hua Y, Wang R, Li D. A Fiber-Based SPR Aptasensor for the In Vitro Detection of Inflammation Biomarkers. Micromachines. 2022; 13(7):1036. https://doi.org/10.3390/mi13071036
Chicago/Turabian StyleHua, Yu, Ridong Wang, and Dachao Li. 2022. "A Fiber-Based SPR Aptasensor for the In Vitro Detection of Inflammation Biomarkers" Micromachines 13, no. 7: 1036. https://doi.org/10.3390/mi13071036