Transgenic Drosophila to Functionally Validate Fall Armyworm ABCC2 Mutations Conferring Bt Resistance
Abstract
:1. Introduction
2. Results
2.1. Expression of SfABCC2 in the Midgut of D. melanogaster Leads to Susceptibility against Bt Toxins
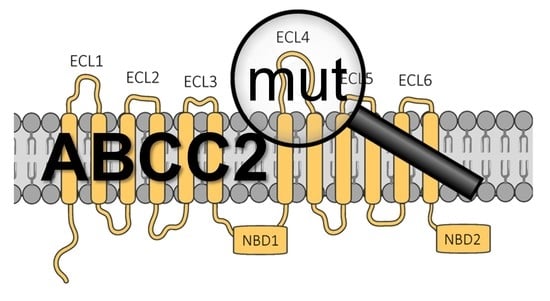
2.2. Mutations in the ABCC2 Transporter Gene Confer Resistance to Xentari in Transgenic Flies
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Drosophila Strains
5.2. Generation of Constructs for Drosophila Transformation
5.3. Generation of Transgenic Flies
5.4. DNA Extraction and PCR Amplification
5.5. Toxicity Bioassays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Overton, K.; Maino, J.L.; Day, R.; Umina, P.A.; Bett, B.; Carnovale, D.; Ekesi, S.; Meagher, R.; Reynolds, O.L. Global Crop Impacts, Yield Losses and Action Thresholds for Fall Armyworm (Spodoptera frugiperda): A Review. Crop Prot. 2021, 145, 105641. [Google Scholar] [CrossRef]
- Sparks, A.N. A Review of the Biology of the Fall Armyworm. Fla. Entomol. 1979, 62, 82–87. [Google Scholar] [CrossRef]
- Tay, W.T.; Meagher, R.L.; Czepak, C.; Groot, A.T. Spodoptera Frugiperda: Ecology, Evolution, and Management Options of an Invasive Species. Annu. Rev. Entomol. 2023, 68, 299–317. [Google Scholar] [CrossRef] [PubMed]
- Kenis, M.; Benelli, G.; Biondi, A.; Calatayud, P.-A.; Day, R.; Desneux, N.; Harrison, R.D.; Kriticos, D.; Rwomushana, I.; van den Berg, J.; et al. Invasiveness, Biology, Ecology, and Management of the Fall Armyworm, Spodoptera Frugiperda. Entomol. Gen. 2023, 43, 187–241. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Y.-P.; Wu, M.-F.; Gao, B.-Y.; Liu, J.; Lee, G.-S.; Otuka, A.; Hu, G. High Risk of the Fall Armyworm Invading Japan and the Korean Peninsula via Overseas Migration. J. Appl. Entomol. 2019, 143, 911–920. [Google Scholar] [CrossRef]
- Tepa-Yotto, G.T.; Tonnang, H.E.Z.; Goergen, G.; Subramanian, S.; Kimathi, E.; Abdel-Rahman, E.M.; Flø, D.; Thunes, K.H.; Fiaboe, K.K.M.; Niassy, S.; et al. Global Habitat Suitability of Spodoptera Frugiperda (JE Smith) (Lepidoptera, Noctuidae): Key Parasitoids Considered for Its Biological Control. Insects 2021, 12, 273. [Google Scholar] [CrossRef]
- Zhou, Y.; Wu, Q.; Zhang, H.; Wu, K. Spread of Invasive Migratory Pest Spodoptera Frugiperda and Management Practices throughout China. J. Integr. Agric. 2021, 20, 637–645. [Google Scholar] [CrossRef]
- Sparks, T.C.; Crossthwaite, A.J.; Nauen, R.; Banba, S.; Cordova, D.; Earley, F.; Ebbinghaus-Kintscher, U.; Fujioka, S.; Hirao, A.; Karmon, D.; et al. Insecticides, Biologics and Nematicides: Updates to IRAC’s Mode of Action Classification—A Tool for Resistance Management. Pestic. Biochem. Physiol. 2020, 167, 104587. [Google Scholar] [CrossRef]
- Adang, M.J.; Crickmore, N.; Jurat-Fuentes, J.L. Chapter Two-Diversity of Bacillus Thuringiensis Crystal Toxins and Mechanism of Action. In Advances in Insect Physiology; Dhadialla, T.S., Gill, S.S., Eds.; Insect Midgut and Insecticidal Proteins; Academic Press: Cambridge, MA, USA, 2014; Volume 47, pp. 39–87. [Google Scholar]
- Bravo, A.; Gill, S.S.; Soberón, M. Mode of Action of Bacillus Thuringiensis Cry and Cyt Toxins and Their Potential for Insect Control. Toxicon 2007, 49, 423–435. [Google Scholar] [CrossRef] [Green Version]
- Heckel, D.G. How Do Toxins from Bacillus Thuringiensis Kill Insects? An Evolutionary Perspective. Arch. Insect Biochem. Physiol. 2020, 104, e21673. [Google Scholar] [CrossRef] [Green Version]
- de Melo, A.L.A.; Soccol, V.T.; Soccol, C.R. Bacillus thuringiensis: Mechanism of Action, Resistance, and New Applications: A Review. Crit. Rev. Biotechnol. 2016, 36, 317–326. [Google Scholar] [CrossRef]
- Jurat-Fuentes, J.L.; Heckel, D.G.; Ferré, J. Mechanisms of Resistance to Insecticidal Proteins from Bacillus Thuringiensis. Annu. Rev. Entomol. 2021, 66, 121–140. [Google Scholar] [CrossRef]
- Endo, H. Molecular and Kinetic Models for Pore Formation of Bacillus Thuringiensis Cry Toxin. Toxins 2022, 14, 433. [Google Scholar] [CrossRef]
- Heckel, D.G. The Essential and Enigmatic Role of ABC Transporters in Bt Resistance of Noctuids and Other Insect Pests of Agriculture. Insects 2021, 12, 389. [Google Scholar] [CrossRef]
- Sato, R.; Adegawa, S.; Li, X.; Tanaka, S.; Endo, H. Function and Role of ATP-Binding Cassette Transporters as Receptors for 3D-Cry Toxins. Toxins 2019, 11, 124. [Google Scholar] [CrossRef] [Green Version]
- Gahan, L.J.; Pauchet, Y.; Vogel, H.; Heckel, D.G. An ABC Transporter Mutation Is Correlated with Insect Resistance to Bacillus Thuringiensis Cry1Ac Toxin. PLoS Genet. 2010, 6, e1001248. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.; Yang, Y.; Shan, Y.; Chakrabarty, S.; Cheng, Y.; Soberón, M.; Bravo, A.; Liu, K.; Wu, K.; Xiao, Y. Two ABC Transporters Are Differentially Involved in the Toxicity of Two Bacillus Thuringiensis Cry1 Toxins to the Invasive Crop-Pest Spodoptera Frugiperda (J. E. Smith). Pest Manag. Sci. 2021, 77, 1492–1501. [Google Scholar] [CrossRef]
- Banerjee, R.; Hasler, J.; Meagher, R.; Nagoshi, R.; Hietala, L.; Huang, F.; Narva, K.; Jurat-Fuentes, J.L. Mechanism and DNA-Based Detection of Field-Evolved Resistance to Transgenic Bt Corn in Fall Armyworm (Spodoptera frugiperda). Sci. Rep. 2017, 7, 10877. [Google Scholar] [CrossRef] [Green Version]
- Flagel, L.; Lee, Y.W.; Wanjugi, H.; Swarup, S.; Brown, A.; Wang, J.; Kraft, E.; Greenplate, J.; Simmons, J.; Adams, N.; et al. Mutational Disruption of the ABCC2 Gene in Fall Armyworm, Spodoptera Frugiperda, Confers Resistance to the Cry1Fa and Cry1A.105 Insecticidal Proteins. Sci. Rep. 2018, 8, 7255. [Google Scholar] [CrossRef] [Green Version]
- Boaventura, D.; Ulrich, J.; Lueke, B.; Bolzan, A.; Okuma, D.; Gutbrod, O.; Geibel, S.; Zeng, Q.; Dourado, P.M.; Martinelli, S.; et al. Molecular Characterization of Cry1F Resistance in Fall Armyworm, Spodoptera Frugiperda from Brazil. Insect Biochem. Mol. Biol. 2020, 116, 103280. [Google Scholar] [CrossRef]
- Franz, L.; Raming, K.; Nauen, R. Recombinant Expression of ABCC2 Variants Confirms the Importance of Mutations in Extracellular Loop 4 for Cry1F Resistance in Fall Armyworm. Toxins 2022, 14, 157. [Google Scholar] [CrossRef] [PubMed]
- Stevens, T.; Song, S.; Bruning, J.B.; Choo, A.; Baxter, S.W. Expressing a Moth Abcc2 Gene in Transgenic Drosophila Causes Susceptibility to Bt Cry1Ac without Requiring a Cadherin-like Protein Receptor. Insect Biochem. Mol. Biol. 2017, 80, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Obata, F.; Tanaka, S.; Kashio, S.; Tsujimura, H.; Sato, R.; Miura, M. Induction of Rapid and Selective Cell Necrosis in Drosophila Using Bacillus Thuringiensis Cry Toxin and Its Silkworm Receptor. BMC Biol. 2015, 13, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denecke, S.; Swevers, L.; Douris, V.; Vontas, J. How Do Oral Insecticidal Compounds Cross the Insect Midgut Epithelium? Insect Biochem. Mol. Biol. 2018, 103, 22–35. [Google Scholar] [CrossRef]
- Horikoshi, R.J.; Bernardi, O.; de Amaral, F.S.A.; Miraldo, L.L.; Durigan, M.R.; Bernardi, D.; Silva, S.S.; Omoto, C. Lack of Relevant Cross-Resistance to Bt Insecticide XenTari in Strains of Spodoptera Frugiperda (J. E. Smith) Resistant to Bt Maize. J. Invertebr. Pathol. 2019, 161, 1–6. [Google Scholar] [CrossRef]
- Douris, V.; Denecke, S.; Van Leeuwen, T.; Bass, C.; Nauen, R.; Vontas, J. Using CRISPR/Cas9 Genome Modification to Understand the Genetic Basis of Insecticide Resistance: Drosophila and Beyond. Pestic. Biochem. Physiol. 2020, 167, 104595. [Google Scholar] [CrossRef]
- Samantsidis, G.-R.; Panteleri, R.; Denecke, S.; Kounadi, S.; Christou, I.; Nauen, R.; Douris, V.; Vontas, J. “What I Cannot Create, I Do Not Understand”: Functionally Validated Synergism of Metabolic and Target Site Insecticide Resistance. Proc. Biol. Sci. 2020, 287, 20200838. [Google Scholar] [CrossRef]
- Wang, J.; Wang, H.; Liu, S.; Liu, L.; Tay, W.T.; Walsh, T.K.; Yang, Y.; Wu, Y. CRISPR/Cas9 Mediated Genome Editing of Helicoverpa Armigera with Mutations of an ABC Transporter Gene HaABCA2 Confers Resistance to Bacillus Thuringiensis Cry2A Toxins. Insect Biochem. Mol. Biol. 2017, 87, 147–153. [Google Scholar] [CrossRef]
- Chung, H.; Bogwitz, M.R.; McCart, C.; Andrianopoulos, A.; ffrench-Constant, R.H.; Batterham, P.; Daborn, P.J. Cis-Regulatory Elements in the Accord Retrotransposon Result in Tissue-Specific Expression of the Drosophila Melanogaster Insecticide Resistance Gene Cyp6g1. Genetics 2007, 175, 1071–1077. [Google Scholar] [CrossRef] [Green Version]
- Douris, V.; Steinbach, D.; Panteleri, R.; Livadaras, I.; Pickett, J.A.; Van Leeuwen, T.; Nauen, R.; Vontas, J. Resistance Mutation Conserved between Insects and Mites Unravels the Benzoylurea Insecticide Mode of Action on Chitin Biosynthesis. Proc. Natl. Acad. Sci. USA 2016, 113, 14692–14697. [Google Scholar] [CrossRef] [Green Version]


| Strain | LC50 (mg/L) | 95% CI a | Slope (±SE) | RR b |
|---|---|---|---|---|
| VK13 × HR-GAL4 (no ABCC2) | >1000 | |||
| ABCC2 × HR-GAL4 (wildtype) | 9.4 | 7.3–11.3 | 6.07 (1.24) | 1 |
| GYdel × HR-GAL4 (mutant) | 27.1 | 24.1–30.2 | 5.35 (0.69) | 2.88 |
| P799K × HR-GAL4 (mutant) | 44.1 | 32.7–51.6 | 6.01 (1.03) | 4.69 |
| Combined × HR-GAL4 (mutant) | 143 | 127–171 | 5.22 (1.12) | 15.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panteleri, R.; Anthousi, A.; Denecke, S.; Boaventura, D.; Nauen, R.; Vontas, J. Transgenic Drosophila to Functionally Validate Fall Armyworm ABCC2 Mutations Conferring Bt Resistance. Toxins 2023, 15, 386. https://doi.org/10.3390/toxins15060386
Panteleri R, Anthousi A, Denecke S, Boaventura D, Nauen R, Vontas J. Transgenic Drosophila to Functionally Validate Fall Armyworm ABCC2 Mutations Conferring Bt Resistance. Toxins. 2023; 15(6):386. https://doi.org/10.3390/toxins15060386
Chicago/Turabian StylePanteleri, Rafaela, Amalia Anthousi, Shane Denecke, Debora Boaventura, Ralf Nauen, and John Vontas. 2023. "Transgenic Drosophila to Functionally Validate Fall Armyworm ABCC2 Mutations Conferring Bt Resistance" Toxins 15, no. 6: 386. https://doi.org/10.3390/toxins15060386