Discovery of a High-Efficient Algicidal Bacterium against Microcystis aeruginosa Based on Examinations toward Culture Strains and Natural Bloom Samples
Abstract
:1. Introduction
2. Results
2.1. Isolation and Identification of Algicidal Bacteria
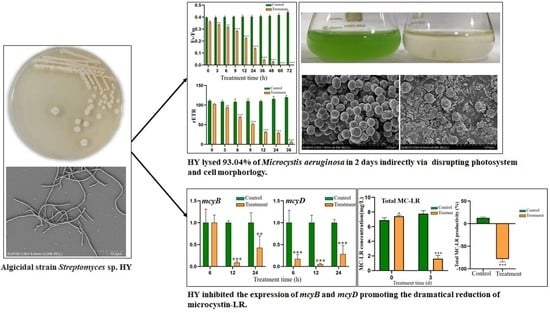
2.2. Algicidal Characteristics of Streptomyces sp. HY
2.3. Algicidal Spectrum of HY
2.4. Effect of HY on Cell Morphology of M. aeruginosa
2.5. Effect of HY Filtrate on the Content of ROS and MDA and the Activity of SOD
2.6. Effect of HY Filtrate on the Photosynthesis System of M. aeruginosa
2.7. Effect of HY Culture on the Content and Distribution of MC-LR
2.8. Effect of HY Filtrate on the Transcription of Key Genes in M. aeruginosa
2.9. Effect of HY Filtrate on Colonies of M. aeruginosa from Natural Bloom Samples
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Algal Strains and Culture Conditions
5.2. Screening and Identification of Algicidal Bacteria
5.3. Determination of Algicidal Mode, Activity and Spectrum
5.4. Determination of Chlorophyll a Concentration
5.5. Determination of Fv/Fm and ETRmax
5.6. Analyses of Intracellular ROS Level
5.7. Content of MDA and Activity of SOD
5.8. Observation of the Cellular Morphology by Scanning Electron Microscope (SEM)
5.9. Analysis of MC-LR
5.10. Effect of HY on Filed Blooms Dominated by Microcystis spp.
5.11. Effect of Strain HY on the Key Gene Expression of M. aeruginosa
5.12. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huisman, J.; Codd, G.A.; Paerl, H.W.; Ibelings, B.W.; Verspagen, J.M.H.; Visser, P.M. Cyanobacterial blooms. Nat. Rev. Microbiol. 2018, 16, 471–483. [Google Scholar] [CrossRef]
- Huo, D.; Gan, N.; Geng, R.; Cao, Q.; Song, L.; Yu, G.; Li, R. Cyanobacterial blooms in China: Diversity, distribution, and cyanotoxins. Harmful Algae 2021, 109, 102106. [Google Scholar] [CrossRef] [PubMed]
- Bouaïcha, N.; Miles, C.O.; Beach, D.G.; Labidi, Z.; Djabri, A.; Benayache, N.Y.; Nguyen-Quang, T. Structural Diversity, Characterization and Toxicology of Microcystins. Toxins 2019, 11, 714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Xie, P. Mechanisms of Microcystin-induced Cytotoxicity and Apoptosis. Mini-Rev. Med. Chem. 2016, 16, 1018–1031. [Google Scholar] [CrossRef]
- McLellan, N.L.; Manderville, R.A. Toxic mechanisms of microcystins in mammals. Toxicol. Res. 2017, 6, 391–405. [Google Scholar] [CrossRef] [Green Version]
- World Health Organization. Guidelines for Drinking-Water Quality: Volume 1, Recommendations. Guidel. Drink. Water Qual. 2004, 38, 104–108. [Google Scholar]
- Paerl, H.W.; Otten, T.G. Harmful Cyanobacterial Blooms: Causes, Consequences, and Controls. Microb. Ecol. 2013, 65, 995–1010. [Google Scholar] [CrossRef]
- Zhou, S.; Shao, Y.; Gao, N.; Deng, Y.; Qiao, J.; Ou, H.; Deng, J. Effects of different algaecides on the photosynthetic capacity, cell integrity and microcystin-LR release of Microcystis aeruginosa. Sci. Total. Environ. 2013, 463–464, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, Z.; Chen, H.; Wen, Y. Effect of hydrogen peroxide on Microcystic aeruginosa: Role of cytochromes P450. Sci. Total. Environ. 2018, 626, 211–218. [Google Scholar] [CrossRef]
- Coyne, K.J.; Wang, Y.; Johnson, G. Algicidal Bacteria: A Review of Current Knowledge and Applications to Control Harmful Algal Blooms. Front. Microbiol. 2022, 13, 871177. [Google Scholar] [CrossRef]
- Li, D.; Kang, X.; Chu, L.; Wang, Y.; Song, X.; Zhao, X.; Cao, X. Algicidal mechanism of Raoultella ornithinolytica against Microcystis aeruginosa: Antioxidant response, photosynthetic system damage and microcystin degradation. Environ. Pollut. 2021, 287, 117644. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Xie, X.; Huang, F.; Xiang, L.; Wang, Y.; Han, T.; Massey, I.Y.; Liang, G.; Pu, Y.; Yang, F. Simultaneous Microcystis algicidal and microcystin synthesis inhibition by a red pigment prodigiosin. Environ. Pollut. 2020, 256, 113444. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ai, H.; Kang, L.; Sun, X.; He, Q. Simultaneous Microcystis Algicidal and Microcystin Degrading Capability by a Single Acinetobacter Bacterial Strain. Environ. Sci. Technol. 2016, 50, 11903–11911. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Hou, X.; Wu, D.; Chang, W.; Zhang, X.; Dai, X.; Du, H.; Zhang, X.; Igarashi, Y.; Luo, F. The characteristics and algicidal mechanisms of cyanobactericidal bacteria, a review. World J. Microbiol. Biotechnol. 2020, 36, 188. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, C.; Chi, Y.; Wu, D.; Dai, X.; Zhang, X.; Igarashi, Y.; Luo, F. Algicidal characterization and mechanism of Bacillus licheniformis Sp34 against Microcystis aeruginosa in Dianchi Lake. J. Basic Microbiol. 2019, 59, 1112–1124. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, M.; Wu, W.; Shi, L.; Luo, L.; Li, P. Differential sensitivity of colonial and unicellular Microcystis strains to an algicidal bacterium Pseudomonas aeruginosa. J. Plankton Res. 2013, 35, 1172–1176. [Google Scholar] [CrossRef]
- Guo, X.; Liu, X.; Wu, L.; Pan, J.; Yang, H. The algicidal activity of Aeromonas sp. strain GLY-2107 against bloom-forming Microcystis aeruginosa is regulated by N-acyl homoserine lactone-mediated quorum sensing. Environ. Microbiol. 2016, 18, 3867–3883. [Google Scholar] [CrossRef]
- Zhang, B.-H.; Ding, Z.-G.; Li, H.-Q.; Mou, X.-Z.; Zhang, Y.-Q.; Yang, J.-Y.; Zhou, E.-M.; Li, W.-J. Algicidal Activity of Streptomyces eurocidicus JXJ-0089 Metabolites and Their Effects on Microcystis Physiology. Appl. Environ. Microbiol. 2016, 82, 5132–5143. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Wang, Y.; Tang, S.; Liang, J.; Lin, W.; Luo, L. Isolation and Identification of Algicidal Compound from Streptomyces and Algicidal Mechanism to Microcystis aeruginosa. PLoS ONE 2013, 8, e76444. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Sun, P.; Zhang, J.; Esquivel-Elizondo, S.; Wu, Y. Microorganisms-based methods for harmful algal blooms control: A review. Bioresour. Technol. 2018, 248, 12–20. [Google Scholar] [CrossRef]
- van Le, V.; Ko, S.-R.; Kang, M.; Lee, S.-A.; Oh, H.-M.; Ahn, C.-Y. Algicide capacity of Paucibacter aquatile DH15 on Microcystis aeruginosa by attachment and non-attachment effects. Environ. Pollut. 2022, 302, 119079. [Google Scholar] [CrossRef]
- Zuo, Y.; Li, L.; Zhang, T.; Zheng, L.; Dai, G.; Liu, L.; Song, L. Contribution of Streptomyces in sediment to earthy odor in the overlying water in Xionghe Reservoir, China. Water Res. 2010, 44, 6085–6094. [Google Scholar] [CrossRef]
- Yu, Y.; Zeng, Y.; Li, J.; Yang, C.; Zhang, X.; Luo, F.; Dai, X. An algicidal Streptomyces amritsarensis strain against Microcystis aeruginosa strongly inhibits microcystin synthesis simultaneously. Sci. Total. Environ. 2019, 650, 34–43. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, J.; Yang, C.; Ding, M.; Hamilton, P.B.; Zhang, X.; Yang, C.; Zhnag, L.; Dai, X. A Streptomyces globisporus strain kills Microcystis aeruginosa via cell-to-cell contact. Sci. Total. Environ. 2021, 769, 144489. [Google Scholar] [CrossRef]
- Paerl, H.W.; Otten, T.G. Blooms Bite the Hand That Feeds Them. Science 2013, 342, 433–434. [Google Scholar] [CrossRef]
- Malanga, G.; Giannuzzi, L.; Hernando, M. The possible role of microcystin (D-Leu1 MC-LR) as an antioxidant on Microcystis aeruginosa (Cyanophyceae). In vitro and in vivo evidence. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2019, 225, 108575. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-H.; Sang, M.; Hwang, S.-J.; Han, M.-S. In situ bacterial mitigation of the toxic cyanobacterium Microcystis aeruginosa: implications for biological bloom control. Limnol. Oceanogr. Methods 2008, 6, 513–522. [Google Scholar] [CrossRef]
- Shao, J.; Jiang, Y.; Wang, Z.; Peng, L.; Luo, S.; Gu, J.; Li, R. Interactions between algicidal bacteria and the cyanobacterium Microcystis aeruginosa: Lytic characteristics and physiological responses in the cyanobacteria. Int. J. Environ. Sci. Technol. 2014, 11, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-D.; Zhu, Y.; Xin, J.-P.; Zhao, C.; Tian, R.-N. Succinic acid inhibits photosynthesis of Microcystis aeruginosa via damaging PSII oxygen-evolving complex and reaction center. Environ. Sci. Pollut. Res. Int. 2021, 28, 58470–58479. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, Y.; Allakhverdiev, S.I.; Murata, N. A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim. Biophys. Acta 2006, 1757, 742–749. [Google Scholar] [CrossRef] [Green Version]
- Chorus, I. Toxic Cyanobacteria in Water—A guide to Their Public Health Consequences, Monitoring and Management; CRC Press: Boca Raton, FL, USA, 1999. [Google Scholar]
- Lee, C.; Jeon, M.S.; Vo, T.-T.; Park, C.; Choi, J.-S.; Kwon, J.; Roh, S.W.; Choi, Y.-E. Establishment of a new strategy against Microcystis bloom using newly isolated lytic and toxin-degrading bacteria. J. Appl. Phycol. 2018, 30, 1795–1806. [Google Scholar] [CrossRef]
- Jiang, Y.; Shao, J.; Wu, X.; Xu, Y.; Li, R. Active and silent members in the mlr gene cluster of a microcystin-degrading bacterium isolated from Lake Taihu, China. FEMS Microbiol. Lett. 2011, 322, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Richards, F.A.; Thompson, T.G. The estimation and characterization of plankton populations by pigment analysis. ii. a spectrophotometric method for the estimation of plankton pigments. J. Mar. Res. 1952, 11, 156–172. [Google Scholar]
- Lu, J.; Zhang, H.; Pan, L.; Guan, W.; Lou, Y. Environmentally relevant concentrations of triclosan exposure promote the horizontal transfer of antibiotic resistance genes mediated by Edwardsiella piscicida. Environ. Sci. Pollut. Res Int. 2022, 29, 64622–64632. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, S.; Wang, Y.; Xu, X. Functional Overlap of hetP and hetZ in Regulation of Heterocyst Differentiation in Anabaena sp. Strain PCC 7120. J. Bacteriol. 2018, 200, 707–717. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]







| Strain | Algicidal Ratio (%) |
|---|---|
| Microcystis aeruginosa FACHB 526 | 99.78 ± 0.00 |
| Microcystis aeruginosa PCC 7806 | 95.20 ± 0.01 |
| Synechocystis sp. PCC 6803 | 88.74 ± 0.01 |
| Pseudoanabaena sp. WZU 1801 | 88.44 ± 0.00 |
| Dolichospermum sp. WZU 1811 | 97.50 ± 0.00 |
| Anabaena sp. PCC 7120 | 64.97 ± 0.06 |
| Scenedesmus obliquuso | 25.74 ± 0.02 |
| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| psbA | GATCCAAGGACGCATTCCTA | CAACGGTGGTCCTTACCAG |
| psbD1 | TCGCAGTGACCATGGAGTAG | TTGAAGACGGTGAAGGTT |
| rbcL | CGTTTCCCCGTCGCTTT | CCGAGTTTGGGTTTGATGGT |
| prx | GCGAATTTAGCAGTATCAACACC | GCGGTGCTGATTTCTTTTTTC |
| sod | GAACCAACCAAGCCCAACC | CAACAATGCCGCCCAAG |
| grpE | CGCAAACGCACAGCCAAGGAA | GTGAATACCCATCTCGCCATC |
| mcyB | CCTACCGAGCGCTTGGG | GAAAATCCCCTAAAGATTCCTGAGT |
| mcyD | ACCCGGAACGGTCATAAATTGG | CGGCTAATCTCTCCAAAACATTGC |
| recA | GCCGAACAAACTAACGTGGT | GGAACCGAACCCATGTC |
| ATP | GTATGGATATCGTGGACACCG | AGATCGACTAATTTGGGAGCG |
| 16S | GGACGGGTGAGTAACGCGTA | CCCATTGCGGAAAATTCCCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Xie, Y.; Zhang, R.; Zhang, Z.; Hu, X.; Cheng, Y.; Geng, R.; Ma, Z.; Li, R. Discovery of a High-Efficient Algicidal Bacterium against Microcystis aeruginosa Based on Examinations toward Culture Strains and Natural Bloom Samples. Toxins 2023, 15, 220. https://doi.org/10.3390/toxins15030220
Zhang H, Xie Y, Zhang R, Zhang Z, Hu X, Cheng Y, Geng R, Ma Z, Li R. Discovery of a High-Efficient Algicidal Bacterium against Microcystis aeruginosa Based on Examinations toward Culture Strains and Natural Bloom Samples. Toxins. 2023; 15(3):220. https://doi.org/10.3390/toxins15030220
Chicago/Turabian StyleZhang, He, Yan Xie, Rongzhen Zhang, Zhongliang Zhang, Xinglong Hu, Yao Cheng, Ruozhen Geng, Zengling Ma, and Renhui Li. 2023. "Discovery of a High-Efficient Algicidal Bacterium against Microcystis aeruginosa Based on Examinations toward Culture Strains and Natural Bloom Samples" Toxins 15, no. 3: 220. https://doi.org/10.3390/toxins15030220