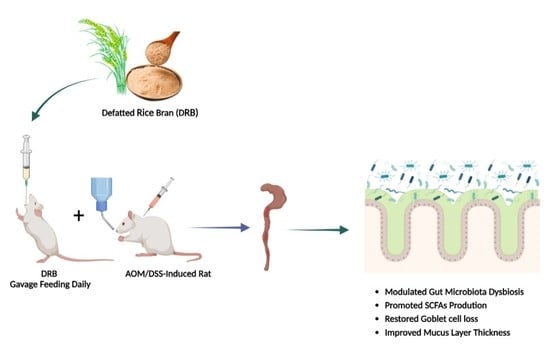
Role of Dietary Defatted Rice Bran in the Modulation of Gut Microbiota in AOM/DSS-Induced Colitis-Associated Colorectal Cancer Rat Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Defatted Rice Bran (DRB)
2.2. Animal Experiment and Sample Collection
2.3. Colonic Goblet Cells and Mucus Layer Thickness Evaluation
2.4. Gut Microbiota Analysis
2.4.1. DNA Extraction
2.4.2. 16S rRNA Amplicon Library Preparation and Sequencing
2.4.3. Bioinformatics Analysis
2.5. Short-Chain Fatty Acid (SCFA) Analysis
2.6. Statistical Analysis
3. Results
3.1. DRB Supplementation Protection against Goblet Cell Loss in AOM/DSS-Induced Colitis-Associated CRC Rats
3.2. DRB Supplementation Restored Intestinal Barrier in AOM/DSS-Induced Colitis-Associated CRC Rats
3.3. DRB Supplementation Modulated the Composition of Gut Microbiota in AOM/DSS-Induced Colitis-Associated CRC Rats
3.3.1. Alpha Diversity
3.3.2. Beta Diversity
3.3.3. Bacterial Taxonomic Composition
3.3.4. Bacterial Biomarkers
3.4. DRB Supplementation Increased Cecal Short-Chain Fatty Acids (SCFAs) Production in AOM/DSS-Induced Colitis-Associated CRC Rats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef]
- Sun, J.; Kato, I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016, 3, 130–143. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.; Fang, L.; Lee, M.H. Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. 2018, 6, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nistal, E.; Fernández-Fernández, N.; Vivas, S.; Olcoz, J.L. Factors Determining Colorectal Cancer: The Role of the Intestinal Microbiota. Front. Oncol. 2015, 5, 220. [Google Scholar] [CrossRef] [Green Version]
- Janney, A.; Powrie, F.; Mann, E.H. Host–microbiota maladaptation in colorectal cancer. Nature 2020, 585, 509–517. [Google Scholar] [CrossRef] [PubMed]
- Hevia, A.; Delgado, S.; Sánchez, B.; Margolles, A. Molecular Players Involved in the Interaction Between Beneficial Bacteria and the Immune System. Front. Microbiol. 2015, 6, 1285. [Google Scholar] [CrossRef] [Green Version]
- Sohail, M.; Rakha, A.; Butt, M.S.; Iqbal, M.J.; Rashid, S. Rice bran nutraceutics: A comprehensive review. Crit. Rev. Food Sci. Nutr. 2017, 57, 3771–3780. [Google Scholar] [CrossRef] [PubMed]
- Sapwarobol, S.; Saphyakhajorn, W.; Astina, J. Biological Functions and Activities of Rice Bran as a Functional Ingredient: A Review. Nutr. Metab. Insights 2021, 14, 11786388211058559. [Google Scholar] [CrossRef]
- Zhuang, X.; Yin, T.; Han, W.; Zhang, X. Chapter 10—Nutritional Ingredients and Active Compositions of Defatted Rice Bran. In Rice Bran and Rice Bran Oil; Cheong, L.-Z., Xu, X., Eds.; AOCS Press: Urbana, IL, USA, 2019; pp. 247–270. [Google Scholar]
- So, W.K.; Law, B.M.; Law, P.T.; Chan, C.W.; Chair, S.Y. Current Hypothesis for the Relationship between Dietary Rice Bran Intake, the Intestinal Microbiota and Colorectal Cancer Prevention. Nutrients 2016, 8, 569. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, P.; Du, T.; Pu, G.; Fan, L.; Gao, C.; Niu, P.; Wu, C.; Zhou, W.; Huang, R. Effects of Increasing Levels of Defatted Rice Bran on Intestinal Physical Barrier and Bacteria in Finishing Pigs. Animals 2019, 9, 1039. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Jiang, Z.; Wang, C.; Zhang, Y.; Gong, T.; Wang, F.; Jin, M.; Wang, Y.; Lu, Z. Co-fermented defatted rice bran alters gut microbiota and improves growth performance, antioxidant capacity, immune status and intestinal permeability of finishing pigs. Anim. Nutr. 2022, 11, 413–424. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, J.; Liu, S.; Fan, Y.; Mei, J.; Liu, X.; Wei, T. Positive intervention of insoluble dietary fiber from defatted rice bran on hyperlipidemia in high fat diet fed rats. J. Food Sci. 2021, 86, 3964–3974. [Google Scholar] [CrossRef]
- Hou, H.; Chen, D.; Zhang, K.; Zhang, W.; Liu, T.; Wang, S.; Dai, X.; Wang, B.; Zhong, W.; Cao, H. Gut microbiota-derived short-chain fatty acids and colorectal cancer: Ready for clinical translation? Cancer Lett. 2022, 526, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Śliżewska, K.; Markowiak-Kopeć, P.; Śliżewska, W. The Role of Probiotics in Cancer Prevention. Cancers 2020, 13, 20. [Google Scholar] [CrossRef]
- Belcheva, A.; Irrazabal, T.; Martin, A. Gut microbial metabolism and colon cancer: Can manipulations of the microbiota be useful in the management of gastrointestinal health? Bioessays 2015, 37, 403–412. [Google Scholar] [CrossRef]
- Li, M.; van Esch, B.C.A.M.; Wagenaar, G.T.M.; Garssen, J.; Folkerts, G.; Henricks, P.A.J. Pro-and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur. J. Pharmacol. 2018, 831, 52–59. [Google Scholar] [CrossRef]
- Mirzaei, R.; Afaghi, A.; Babakhani, S.; Sohrabi, M.R.; Hosseini-Fard, S.R.; Babolhavaeji, K.; Khani Ali Akbari, S.; Yousefimashouf, R.; Karampoor, S. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021, 139, 111619. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Lazarova, D.L.; Bordonaro, M. Mechanisms linking dietary fiber, gut microbiota and colon cancer prevention. World J. Gastrointest. Oncol. 2014, 6, 41–51. [Google Scholar] [CrossRef]
- Liu, H.; Wang, J.; He, T.; Becker, S.; Zhang, G.; Li, D.; Ma, X. Butyrate: A Double-Edged Sword for Health? Adv. Nutr. 2018, 9, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Sheflin, A.M.; Borresen, E.C.; Wdowik, M.J.; Rao, S.; Brown, R.J.; Heuberger, A.L.; Broeckling, C.D.; Weir, T.L.; Ryan, E.P. Pilot dietary intervention with heat-stabilized rice bran modulates stool microbiota and metabolites in healthy adults. Nutrients 2015, 7, 1282–1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, D.G.; Borresen, E.C.; Brown, R.J.; Ryan, E.P. Heat-stabilised rice bran consumption by colorectal cancer survivors modulates stool metabolite profiles and metabolic networks: A randomised controlled trial. Br. J. Nutr. 2017, 117, 1244–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nair, A.B.; Jacob, S. A simple practice guide for dose conversion between animals and human. J. Basic Clin. Pharm. 2016, 7, 27–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribeiro, W.R.; Vinolo, M.A.R.; Calixto, L.A.; Ferreira, C.M. Use of Gas Chromatography to Quantify Short Chain Fatty Acids in the Serum, Colonic Luminal Content and Feces of Mice. Bio-Protocol 2018, 8, e3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tajasuwan, L.; Kettawan, A.; Rungruang, T.; Wunjuntuk, K.; Prombutara, P.; Muangnoi, C.; Kettawan, A.K. Inhibitory Effect of Dietary Defatted Rice Bran in an AOM/DSS-Induced Colitis-Associated Colorectal Cancer Experimental Animal Model. Foods 2022, 11, 3488. [Google Scholar] [CrossRef] [PubMed]
- Koh, A.; De Vadder, F.; Kovatcheva-Datchary, P.; Bäckhed, F. From Dietary Fiber to Host Physiology: Short-Chain Fatty Acids as Key Bacterial Metabolites. Cell 2016, 165, 1332–1345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagpal, R.; Wang, S.; Solberg Woods, L.C.; Seshie, O.; Chung, S.T.; Shively, C.A.; Register, T.C.; Craft, S.; McClain, D.A.; Yadav, H. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-human Primate, and Human Feces. Front. Microbiol. 2018, 9, 2897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD—What role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. Biomed. Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.G.; Lee, Y.-R.; Lee, A.R.; Park, C.H.; Han, D.S.; Eun, C.S. Role of the global gut microbial community in the development of colitis-associated cancer in a murine model. Biomed. Pharmacother. 2021, 135, 111206. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef] [Green Version]
- Umar, S. Citrobacter Infection and Wnt signaling. Curr. Color. Cancer Rep. 2012, 8, 298–306. [Google Scholar] [CrossRef] [Green Version]
- Keku, T.O.; Dulal, S.; Deveaux, A.; Jovov, B.; Han, X. The gastrointestinal microbiota and colorectal cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G351–G363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.; Kim, N.-E.; Yoon, H.; Shin, C.M.; Kim, N.; Lee, D.H.; Park, J.Y.; Choi, C.H.; Kim, J.G.; Kim, Y.-K.; et al. Fecal Microbiota and Gut Microbe-Derived Extracellular Vesicles in Colorectal Cancer. Front. Oncol. 2021, 11, 650026. [Google Scholar] [CrossRef]
- Pierce, E.S. Could Mycobacterium avium subspecies paratuberculosis cause Crohn’s disease, ulcerative colitis…and colorectal cancer? Infect. Agent. Cancer 2018, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Yang, Y.; Sun, B.; Jing, Y.; Deng, W. Dietary Fiber, Gut Microbiota, Short-Chain Fatty Acids, and Host Metabolism. Am. J. Life Sci. 2021, 9, 162–172. [Google Scholar] [CrossRef]
- Abreu, Y.A.A.T.; Milke-García, M.P.; Argüello-Arévalo, G.A.; Calderón-de la Barca, A.M.; Carmona-Sánchez, R.I.; Consuelo-Sánchez, A.; Coss-Adame, E.; García-Cedillo, M.F.; Hernández-Rosiles, V.; Icaza-Chávez, M.E.; et al. Dietary fiber and the microbiota: A narrative review by a group of experts from the Asociación Mexicana de Gastroenterología. Rev. Gastroenterol. Mex. 2021, 86, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Zhang, X.; Luo, T.; Wang, D.; Sun, Y.; Dai, J. Effects of Short-Term Dietary Fiber Intervention on Gut Microbiota in Young Healthy People. Diabetes Metab. Syndr. Obes. 2021, 14, 3507–3516. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, B.; Cong, W.; Zhang, M.; Li, Z.; Li, Y.; Liang, S.; Chen, K.; Yang, D.; Wu, Z. Amelioration of AOM/DSS-Induced Murine Colitis-Associated Cancer by Evodiamine Intervention is Primarily Associated with Gut Microbiota-Metabolism-Inflammatory Signaling Axis. Front. Pharmacol. 2021, 12, 797605. [Google Scholar] [CrossRef]
- Geirnaert, A.; Calatayud, M.; Grootaert, C.; Laukens, D.; Devriese, S.; Smagghe, G.; De Vos, M.; Boon, N.; Van de Wiele, T. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017, 7, 11450. [Google Scholar] [CrossRef] [Green Version]
- Tamanai-Shacoori, Z.; Smida, I.; Bousarghin, L.; Loreal, O.; Meuric, V.; Fong, S.B.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Roseburia spp.: A marker of health? Future Microbiol. 2017, 12, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Sivamaruthi, B.S.; Kesika, P.; Chaiyasut, C. The Role of Probiotics in Colorectal Cancer Management. Evid. Based Complement. Alternat. Med. 2020, 2020, 3535982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parada Venegas, D.; De la Fuente, M.K.; Landskron, G.; González, M.J.; Quera, R.; Dijkstra, G.; Harmsen, H.J.M.; Faber, K.N.; Hermoso, M.A. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 2019, 10, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geerlings, S.Y.; Kostopoulos, I.; de Vos, W.M.; Belzer, C. Akkermansia muciniphila in the Human Gastrointestinal Tract: When, Where, and How? Microorganisms 2018, 6, 75. [Google Scholar] [CrossRef] [Green Version]
- Plamada, D.; Vodnar, D.C. Polyphenols-Gut Microbiota Interrelationship: A Transition to a New Generation of Prebiotics. Nutrients 2021, 14, 137. [Google Scholar] [CrossRef]
- Schneeberger, M.; Everard, A.; Gómez-Valadés, A.G.; Matamoros, S.; Ramírez, S.; Delzenne, N.M.; Gomis, R.; Claret, M.; Cani, P.D. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci. Rep. 2015, 5, 16643. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Cai, K.; Xiao, Q.; He, L.; Xie, L.; Liu, Z. Akkermansia muciniphila administration exacerbated the development of colitis-associated colorectal cancer in mice. J. Cancer 2022, 13, 124–133. [Google Scholar] [CrossRef]
- Nealon, N.J.; Parker, K.D.; Lahaie, P.; Ibrahim, H.; Maurya, A.K.; Raina, K.; Ryan, E.P. Bifidobacterium longum-fermented rice bran and rice bran supplementation affects the gut microbiome and metabolome. Benef. Microbes 2019, 10, 823–839. [Google Scholar] [CrossRef]
- Parker, K.D.; Maurya, A.K.; Ibrahim, H.; Rao, S.; Hove, P.R.; Kumar, D.; Kant, R.; Raina, B.; Agarwal, R.; Kuhn, K.A.; et al. Dietary Rice Bran-Modified Human Gut Microbial Consortia Confers Protection against Colon Carcinogenesis Following Fecal Transfaunation. Biomedicines 2021, 9, 144. [Google Scholar] [CrossRef]
- Zhao, G.; Zhang, R.; Huang, F.; Dong, L.; Liu, L.; Jia, X.; Chi, J.; Ma, Y.; Deng, M.; Chen, Y.; et al. Hydrolyzed Bound Phenolics from Rice Bran Alleviate Hyperlipidemia and Improve Gut Microbiota Dysbiosis in High-Fat-Diet Fed Mice. Nutrients 2022, 14, 1277. [Google Scholar] [CrossRef]
- Komiyama, Y.; Andoh, A.; Fujiwara, D.; Ohmae, H.; Araki, Y.; Fujiyama, Y.; Mitsuyama, K.; Kanauchi, O. New prebiotics from rice bran ameliorate inflammation in murine colitis models through the modulation of intestinal homeostasis and the mucosal immune system. Scand. J. Gastroenterol. 2011, 46, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Islam, J.; Koseki, T.; Watanabe, K.; Budijanto, S.; Oikawa, A.; Alauddin, M.; Goto, T.; Aso, H.; Komai, M.; Shirakawa, H. Dietary Supplementation of Fermented Rice Bran Effectively Alleviates Dextran Sodium Sulfate-Induced Colitis in Mice. Nutrients 2017, 9, 747. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.E.; Gustafsson, J.K.; Sjöberg, K.E.; Petersson, J.; Holm, L.; Sjövall, H.; Hansson, G.C. Bacteria penetrate the inner mucus layer before inflammation in the dextran sulfate colitis model. PLoS ONE 2010, 5, e12238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solomon, L.; Mansor, S.; Mallon, P.; Donnelly, E.; Hoper, M.; Loughrey, M.; Kirk, S.; Gardiner, K. The dextran sulphate sodium (DSS) model of colitis: An overview. Comp. Clin. Path. 2010, 19, 235–239. [Google Scholar] [CrossRef]
- Birchenough, G.M.H.; Johansson, M.E.; Gustafsson, J.K.; Bergström, J.H.; Hansson, G.C. New developments in goblet cell mucus secretion and function. Mucosal Immunol. 2015, 8, 712–719. [Google Scholar] [CrossRef] [Green Version]
- Duangnumsawang, Y.; Zentek, J.; Goodarzi Boroojeni, F. Development and Functional Properties of Intestinal Mucus Layer in Poultry. Front. Immunol. 2021, 12, 745849. [Google Scholar] [CrossRef]
- Grondin, J.A.; Kwon, Y.H.; Far, P.M.; Haq, S.; Khan, W.I. Mucins in Intestinal Mucosal Defense and Inflammation: Learning From Clinical and Experimental Studies. Front. Immunol. 2020, 11, 2054. [Google Scholar] [CrossRef]
- Kim, Y.S.; Ho, S.B. Intestinal goblet cells and mucins in health and disease: Recent insights and progress. Curr. Gastroenterol. Rep. 2010, 12, 319–330. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, B.O. Fight them or feed them: How the intestinal mucus layer manages the gut microbiota. Gastroenterol. Rep. 2019, 7, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Paone, P.; Cani, P.D. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut 2020, 69, 2232–2243. [Google Scholar] [CrossRef]
- Fang, J.; Wang, H.; Zhou, Y.; Zhang, H.; Zhou, H.; Zhang, X. Slimy partners: The mucus barrier and gut microbiome in ulcerative colitis. Exp. Mol. Med. 2021, 53, 772–787. [Google Scholar] [CrossRef] [PubMed]
- Desai, M.S.; Seekatz, A.M.; Koropatkin, N.M.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A Dietary Fiber-Deprived Gut Microbiota Degrades the Colonic Mucus Barrier and Enhances Pathogen Susceptibility. Cell 2016, 167, 1339–1353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; He, C.; Liu, Y.; Zhao, H.; Long, L.; Gai, X.; Zhao, H. Soluble dietary fiber protects intestinal mucosal barrier by improving intestinal flora in a murine model of sepsis. Biomed. Pharmacother. 2020, 129, 110343. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, I.A.; Havler, M.E.; Dettmar, P.W.; Allen, A.; Pearson, J.P. Colonic mucus: Secretion and turnover in relation to dietary fibre intake. Proc. Nutr. Soc. 2003, 62, 245–249. [Google Scholar] [CrossRef]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tajasuwan, L.; Kettawan, A.; Rungruang, T.; Wunjuntuk, K.; Prombutara, P. Role of Dietary Defatted Rice Bran in the Modulation of Gut Microbiota in AOM/DSS-Induced Colitis-Associated Colorectal Cancer Rat Model. Nutrients 2023, 15, 1528. https://doi.org/10.3390/nu15061528
Tajasuwan L, Kettawan A, Rungruang T, Wunjuntuk K, Prombutara P. Role of Dietary Defatted Rice Bran in the Modulation of Gut Microbiota in AOM/DSS-Induced Colitis-Associated Colorectal Cancer Rat Model. Nutrients. 2023; 15(6):1528. https://doi.org/10.3390/nu15061528
Chicago/Turabian StyleTajasuwan, Laleewan, Aikkarach Kettawan, Thanaporn Rungruang, Kansuda Wunjuntuk, and Pinidphon Prombutara. 2023. "Role of Dietary Defatted Rice Bran in the Modulation of Gut Microbiota in AOM/DSS-Induced Colitis-Associated Colorectal Cancer Rat Model" Nutrients 15, no. 6: 1528. https://doi.org/10.3390/nu15061528