Early Detection of Wild Rocket Tracheofusariosis Using Hyperspectral Image-Based Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strain
2.2. Experimental Plant Infection
2.3. Genomic DNA Extraction
2.4. In Planta Pathogen Quantification
2.5. Hyperspectral Imaging Workflow
2.6. Hyperspectral Vegetation Indices
2.7. Statistical Analyses and Machine Learning Pipeline
2.7.1. Testing the Differences between Plants’ Spectral Profiles
2.7.2. Machine Learning Pipeline and Models’ Performance Measure Strategy
2.7.3. The Extreme Gradient Boosting Algorithm
3. Results
3.1. FOR Microscopic and Molecular Characterization
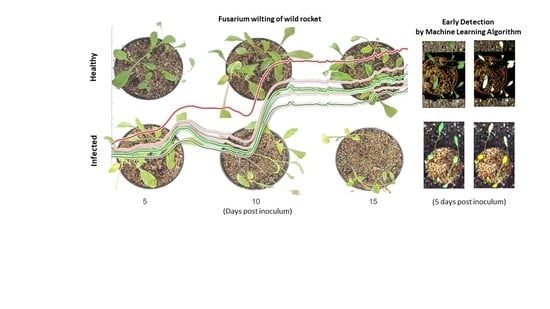
3.2. Progression of Wild Rocket Tracheofusariosis
3.3. Plant Reflectance Datasets
3.4. Temporal Patterns of Hyperspectral Vegetative Indices
3.5. PERMANOVA Results
3.6. Machine Learning Models Results and Early Detection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bonasia, A.; Lazzizera, C.; Elia, A.; Conversa, G. Nutritional, biophysical and physiological characteristics of wild rocket genotypes as affected by soilless cultivation system, salinity level of nutrient solution and growing period. Front. Plant Sci. 2017, 8, 300. [Google Scholar] [CrossRef] [Green Version]
- Caruso, G.; Stoleru, V.; De Pascale, S.; Cozzolino, E.; Pannico, A.; Giordano, M.; Teliban, G.; Cuciniello, A.; Rouphael, Y. Production, leaf quality and antioxidants of perennial wall rocket as affected by crop cycle and mulching type. Agronomy 2019, 9, 194. [Google Scholar] [CrossRef] [Green Version]
- Freshplaza. Il Valore del Settore della Rucola in Italia e’ Stimato in 30–40 Milioni di Euro Solo per L’export 2012. Available online: https://www.freshplaza.it/article/4044942/il-valore-del-settore-della-rucola-in-italia-e-stimato-in-30-40-milioni-di-euro-solo-per-l-export/ (accessed on 12 June 2021).
- Caruso, G.; Parrella, G.; Giorgini, M.; Nicoletti, R. Crop systems, quality and protection of Diplotaxis tenuifolia. Agriculture 2018, 8, 55. [Google Scholar] [CrossRef] [Green Version]
- Nicoletti, R.; Raimo, F.; Miccio, G. Diplotaxis tenuifolia: Biology, production and properties. Eur. J. Plant Sci. Biotechnol. 2007, 1, 36–43. [Google Scholar]
- Li, X.; Zhang, Y.; Ding, C.; Jia, Z.; He, Z.; Zhang, T.; Wang, X. Declined soil suppressiveness to Fusarium oxysporum by rhizosphere microflora of cotton in soil sickness. Biol. Fertil. Soils 2015, 51, 935–946. [Google Scholar] [CrossRef]
- Garibaldi, A.; Gilardi, G.; Gullino, M.L. First of Fusarium oxysporum on Eruca vesicaria and Diplotaxis spp. in Europe. Plant Dis. 2003, 87, 2. [Google Scholar] [CrossRef]
- Garibaldi, A.; Gilardi, G.; Gullino, M.L. Evidence for an expanded host range of Fusarium oxysporum f. sp. raphani. Phytoparasitica 2006, 34, 115–121. [Google Scholar] [CrossRef]
- Catti, A.; Pasquali, M.; Ghiringhelli, D.; Garibaldi, A.; Gullino, M.L. Analysis of vegetative compatibility groups of Fusarium oxysporum from Eruca vesicaria and Diplotaxis tenuifolia. J. Phytopathol. 2007, 155, 61–64. [Google Scholar] [CrossRef]
- Taylor, A.; Barnes, A.; Jackson, A.C.; Clarkson, J.P. First Report of Fusarium oxysporum and Fusarium redolens causing wilting and yellowing of wild rocket (Diplotaxis tenuifolia) in the United Kingdom. Plant Dis. 2019, 103, 6. [Google Scholar] [CrossRef]
- Garibaldi, A.; Gilardi, G.; Pasquali, M.; Kejji, S.; Gullino, M.L. Seed transmission of Fusarium oxysporum of Eruca vesicaria and Diplotaxis muralis. J. Plant Dis. Prot. 2004, 111, 345–350. [Google Scholar]
- Martos, V.; Ahmad, A.; Cartujo, P.; Ordoñez, J. Ensuring agricultural sustainability through remote sensing in the era of agriculture 5.0. Appl. Sci. 2021, 11, 5911. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Zhao, Y.R.; Li, X.; Yu, K.Q.; Cheng, F.; He, Y. Hyperspectral imaging for determining pigment contents in cucumber leaves in response to angular leaf spot disease. Sci. Rep. 2016, 6, 27790. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Experiment. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Behmann, J.; Mahlein, A.K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Appeltans, S.; Pieters, J.G.; Mouazen, A.M. Detection of leek white tip disease under field conditions using hyperspectral proximal sensing and supervised machine learning. Comput. Electron. Agric. 2021, 190, 106453. [Google Scholar] [CrossRef]
- Bauriegel, E.; Herppich, W.B. Hyperspectral and chlorophyll fluorescence imaging for early detection of plant diseases, with special reference to Fusarium spec. infections on Wheat. Agriculture 2014, 4, 32–57. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lee, W.S.; Li, M.Z.; Zheng, L.H.; Ritenour, M.A. Non-destructive recognition and classification of citrus fruit blemishes based on ant colony optimized spectral information. Postharvest Biol. Technol. 2018, 143, 119–128. [Google Scholar] [CrossRef]
- Tsaftaris, S.A.; Minervini, M.; Scharr, H. Machine learning for plant phenotyping needs image processing. Trends Plant Sci. 2016, 21, 989–991. [Google Scholar] [CrossRef] [Green Version]
- Abdulridha, J.; Batuman, O.; Ampatzidis, Y. UAV-based remote sensing technique to detect citrus canker disease utilizing hyperspectral imaging and machine learning. Remote Sens. 2019, 11, 1373. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Chu, B.; Zhang, C.; Liu, F.; Jiang, L.; He, Y. Hyperspectral imaging for presymptomatic detection of tobacco disease with successive projections algorithm and machine-learning classifiers. Sci. Rep. 2017, 7, 4125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Li, X.; Zhu, P.; Xu, N.; He, Y. Hyperspectral reflectance imaging combined with multivariate analysis for diagnosis of Sclerotinia stem rot on Arabidopsis thaliana leaves. Appl. Sci. 2019, 9, 2092. [Google Scholar] [CrossRef] [Green Version]
- Hornero, A.; Zarco-Tejada, P.J.; Quero, J.L.; North, P.R.J.; Ruiz-Gómez, F.J.; Sánchez-Cuesta, R.; Hernandez-Clemente, R. Modelling hyperspectral- and thermal-based plant traits for the early detection of Phytophthora-induced symptoms in oak decline. Remote Sens. Environ. 2021, 263, 112570. [Google Scholar] [CrossRef]
- Nguyen, C.; Sagan, V.; Maimaitiyiming, M.; Maimaitijiang, M.; Bhadra, S.; Kwasniewski, M.T. Early detection of plant viral disease using hyperspectral imaging and deep learning. Sensors 2021, 21, 742. [Google Scholar] [CrossRef]
- Wei, X.; Johnson, M.A.; Langston, D.B.; Mehl, H.L.; Li, S. Identifying optimal wavelengths as disease signatures using hyperspectral sensor and machine learning. Remote Sens. 2021, 13, 2833. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar] [CrossRef] [Green Version]
- Larkin, R.P.; Honeycutt, C.W. Effects of different 3-year cropping systems on soil microbial communities and Rhizoctonia diseases of potato. Phytopathology 2006, 96, 68–79. [Google Scholar] [CrossRef] [Green Version]
- Chiang, K.S.; Liu, H.I.; Bock, C.H.A. Discussion on disease severity index values. Part I: Warning on inherent errors and suggestions to maximize accuracy. Ann. Appl. Biol. 2017, 171, 139–154. [Google Scholar] [CrossRef]
- Manganiello, G.; Nicastro, N.; Caputo, M.; Zaccardelli, M.; Cardi, T.; Pane, C. Functional hyperspectral imaging by high-related vegetation indices to track the wide-spectrum Trichoderma biocontrol activity against soil-borne diseases of baby-leaf vegetables. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Atoui, A.; El Khoury, A.; Kallassy, M.; Lebrihi, A. Quantification of Fusarium graminearum and Fusarium culmorum by real-time PCR system and zearalenone assessment in maize. Int. J. Food Microbiol. 2012, 15, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Khandekar, S.; Leisner, S. Soluble silicon modulates expression of Arabidopsis thaliana genes involved in copper stress. J. Plant Physiol. 2011, 168, 699–705. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Van Etten, J.; Cheng, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.A.; Hiemstra, P.; Hingee, K.; Karney, C.; Mattiuzzi, M.; et al. Package ‘raster’. R Package 2015, 734, 1–94. [Google Scholar]
- Kassambara, A.; Mundt, F. Package ‘Factoextra’. Extract and Visualize the Results of Multivariate Data Analyses 76. 2017. Available online: https://cloud.r-project.org/package=factoextra (accessed on 16 June 2021).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.; Behmann, J.; Steier, A.; Kraska, T.; Muller, O.; Rascher, U.; Mahlein, A.K. Quantitative assessment of disease severity and rating of barley cultivars based on hyperspectral imaging in a non-invasive, automated phenotyping platform. Plant Methods 2018, 14, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, J.R. Remote Sensing of the Environment: An Earth Resource Perspective, 2nd ed.; Pearson Education: London, UK, 2006. [Google Scholar]
- Bravo, C.; Moshou, D.; Oberti, R.; West, J.; McCartney, A.; Bodria, L.; Ramon, H. Foliar disease detection in the field using optical sensor fusion. Agric. Eng. Int. CIGR J. Sci. Res. Dev. 2004, 6, 1–14. [Google Scholar]
- Hillnhütter, C.; Mahlein, A.K.; Sikora, R.A.; Oerke, E.C. Use of imaging spectroscopy to discriminate symptoms caused by Heterodera schachtii and Rhizoctonia solani on sugar beet. Precis. Agric. 2012, 13, 17–32. [Google Scholar] [CrossRef]
- Wahabzada, M.; Kersting, K.; Bauckhage, C.; Römer, C.; Ballvora, A.; Pinto, F.; Rascher, U.; Léon, J.; Plümer, L. Latent Dirichlet Allocation Uncovers Spectral Characteristics of Drought Stressed Plants. In Proceedings of the 28th Conference, Avalon, CA, USA, 15–17 August 2012. [Google Scholar]
- Thomas, S.; Wahabzada, M.; Kuska, M.; Rascher, U.; Mahlein, A.K. Observation of plant–pathogen interaction by simultaneous hyperspectral imaging reflection and transmission measurements. Funct. Plant Biol. 2017, 44, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Kuska, M.; Wahabzada, M.; Leucker, M.; Dehne, H.W.; Kersting, K.; Oerke, E.C.; Steiner, U.; Mahlein, A.K. Hyperspectral phenotyping on the microscopic scale: Towards automated characterization of plant-pathogen interactions. Plant Methods 2015, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Leucker, M.; Mahlein, A.K.; Steiner, U.; Oerke, E.C. Improvement of lesion phenotyping in Cercospora beticola-sugar beet interaction by hyperspectral imaging. Phytopathology 2016, 1, 1–30. [Google Scholar] [CrossRef] [Green Version]
- Hillnhütter, C.; Mahlein, A.K.; Sikora, R.A.; Oerke, E.C. Remote sensing to detect plant stress induced by Heterodera schachtii and Rhizoctonia solani in sugar beet fields. Field Crops Res. 2011, 122, 70–77. [Google Scholar] [CrossRef]
- Susič, N.; Žibrat, U.; Širca, S.; Strajnar, P.; Razinger, J.; Knapič, M.; Vončina, A.; Urek, G.; Gerič Stare, B. Discrimination between abiotic and biotic drought stress in tomatoes using hyperspectral imaging. Sens. Actuators B Chem. 2018, 273, 842–852. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Sun, Y.; Sun, G.; Liu, X.; Zhai, L.; Shen, Q.; Guo, S. Water balance altered in cucumber plants infected with Fusarium oxysporum f. sp. cucumerinum. Sci. Rep. 2015, 5, 7722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pshibytko, N.L.; Zenevich, L.A.; Kabashnikova, L.F. Changes in the photosynthetic apparatus during Fusarium wilt of tomato. Russ. J. Plant Physiol. 2006, 53, 25–31. [Google Scholar] [CrossRef]
- Olivain, C.; Alabouvette, C. Process of tomato root colonization by a pathogenic strain of Fusarium oxysporum f. sp. lycopersici in comparison with a non-pathogenic strain. New Phytol. 1999, 141, 497–510. [Google Scholar] [CrossRef]
- Benhamou, N.; Garand, C. Cytological analysis of defense related mechanisms induced in pea root tissues in response to colonization by nonpathogenic Fusarium oxysporum Fo47. Phytopathology 2001, 91, 730–740. [Google Scholar] [CrossRef] [Green Version]
- Pu, Z.; Ino, Y.; Kimura, Y.; Tago, A.; Shimizu, M.; Natsume, S.; Sano, Y.; Fujimoto, R.; Kaneko, K.; Shea, D.J.; et al. Changes in the proteome of xylem sap in Brassica oleracea in response to Fusarium oxysporum Stress. Front. Plant Sci. 2016, 7, 31. [Google Scholar] [CrossRef] [Green Version]
- Lagopodi, A.L.; Ram, A.F.J.; Lamers, G.E.M.; Punt, P.J.; Van den Hondel, C.A.M.J.J.; Lugtenberg, B.J.J.; Bloemberg, G.V. Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol. Plant Microbe Interact. 2002, 15, 172–179. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Beckman, C.H.; Mueller, W.C. The rate of vascular colonization as a measure of the genotypic interaction between various cultivars of tomato and various formae or races of Fusarium oxysporum. Physiol. Mol. Plant Pat. 1995, 46, 29–43. [Google Scholar] [CrossRef]
- Baayen, R.P.; van der Plas, C.H. Localization ability, latent period and wilting rate in eleven carnation cultivars with partial resistance to Fusarium wilt. Euphytica 1992, 59, 165–174. [Google Scholar] [CrossRef]
- Kang, Z.; Buchenauer, H. Cytology and ultrastructure of the infection of wheat spikes by Fusarium culmorum. Mycol. Res. 2000, 104, 1083–1093. [Google Scholar] [CrossRef]
- Bauriegel, E.; Giebel, A.; Geyer, M.; Schmidt, U.; Herppich, W.B. Early detection of Fusarium infection in wheat using hyper-spectral imaging. Comput. Electron. Agric. 2011, 75, 304–312. [Google Scholar] [CrossRef]
- Muhammed, H.H.; Larsolle, A. Feature vector based analysis of hyperspectral crop reflectance data for discrimination and quantification of fungal disease severity in wheat. Biosyst. Eng. 2003, 86, 125–134. [Google Scholar] [CrossRef]
- Chitarra, W.; Siciliano, I.; Ferrocino, I.; Gullino, M.L.; Garibaldi, A. Effect of elevated atmospheric CO2 and temperature on the disease severity of rocket plants caused by Fusarium Wilt under phytotron conditions. PLoS ONE 2015, 10, e0140769. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Xiong, Y.; Ling, N.; Shen, Q.; Guo, S. Fusaric acid accelerates the senescence of leaf in banana when infected by Fusarium. World J. Microbiol. Biotechnol. 2014, 30, 1399–1408. [Google Scholar] [CrossRef]
- Wang, M.; Ling, N.; Dong, X.; Liu, X.; Shen, Q.; Guo, S. Effect of fusaric acid on the leaf physiology of cucumber seedlings. Eur. J. Plant. Pathol. 2014, 138, 103–112. [Google Scholar] [CrossRef]
- Mendoza-Vargas, L.A.; Villamarín-Romero, W.P.; Cotrino-Tierradentro, A.S.; Ramírez-Gil, J.G.; Chávez-Arias, C.C.; Restrepo-Díaz, H.; Gómez-Caro, S. Physiological response of cape gooseberry plants to Fusarium oxysporum f. sp. physali, fusaric acid, and water deficit in a hydrophonic system. Front. Plant Sci. 2021, 12, 702842. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, H.B.; Upadhyay, R.S. Role of fusaric acid in the development of ‘Fusarium wilt’ symptoms in tomato: Physiological, biochemical and proteomic perspectives. Plant Physiol. Biochem. 2017, 118, 320–332. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, M.; Li, Y.; Gu, Z.; Ling, N.; Shen, Q.; Guo, S. Wilted cucumber plants infected by Fusarium oxysporum f. sp. cucumerinum do not suffer from water shortage. Ann. Bot. 2017, 120, 427–436. [Google Scholar] [CrossRef]
- Ceccato, P.; Gobron, N.; Flasse, S.; Pinty, B.; Tarantola, S. Designing a spectral index to estimate vegetation water content from remote sensing data: Part I: Theoretical approach. Remote Sens. Environ. 2002, 82, 188–197. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Pushnik, J.C.; Dobrowski, S. Steady-state chlorophyll a fluorescence detection from canopy derivative reflectance and double-peak red-edge effects. Remote Sens. Environ. 2003, 84, 283–294. [Google Scholar] [CrossRef]
- Marín Ortiz, J.C.; Hoyos Carvajal, L.M.; Botero Fernandez, V. Detection of significant wavelengths for identifying and classifying during the incubation period and water stress in plants using reflectance spectroscopy. J. Plant Prot. Res. 2019, 59, 244–254. [Google Scholar] [CrossRef]
- Carter, G.A.; Miller, R.L. Early detection of plant stress by digital imaging within narrow stress-sensitive wavebands. Remote Sens. Environ. 1994, 50, 295–302. [Google Scholar] [CrossRef]
- Zhou, G.; Yin, X. Assessing nitrogen nutritional status, biomass and yield of cotton with NDVI, SPAD and petiole sap nitrate concentration. Exp. Agric. 2018, 54, 531–548. [Google Scholar] [CrossRef]
- Steven, M.D.; Jaggard, K.W. Advances in Crop Monitoring by Remote Sensing. In Advances in Environmental Remote Sensing; Danson, F.M., Plummer, S.E., Eds.; John and Wiley and Sons: Hoboken, NJ, USA, 1995; pp. 143–156. [Google Scholar]
- Smith, R.C.G.; Adams, J.; Stephens, D.J.; Hick, P.T. Forecasting wheat yield in a Mediterranean-type environment from the NOAA satellite. Aus. J. Agric. Res. 1995, 46, 113. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Lichtenthaler, H.K. Detection of red edge position and chlorophyll content by reflectance measurements near 700 nm. J. Plant Physiol. 1996, 148, 501–508. [Google Scholar] [CrossRef]
- Tilley, D.R.; Ahmed, M.; Son, J.H.; Badrinarayanan, H. Hyperspectral reflectance of emergent macrophytes as an indicator of water column ammonia in an oligohaline, subtropical marsh. Ecol. Eng. 2003, 21, 153–163. [Google Scholar] [CrossRef]
- Steddom, K.; Heidel, G.; Jones, D.; Rush, C.M. Remote detection of rhizomania in sugar beets. Phytopathology 2003, 93, 720–726. [Google Scholar] [CrossRef] [Green Version]
- Stagakis, S.; González-Dugo, V.; Cid, P.; Guillén-Climent, M.L.; Zarco-Tejada, P.J. Monitoring water stress and fruit quality in an orange orchard under regulated deficit irrigation using narrow-band structural and physiological remote sensing indices. ISPRS J. Photogramm. Remote Sens. 2012, 71, 47–61. [Google Scholar] [CrossRef] [Green Version]
- Sandmann, M.; Grosch, R.; Graefe, J. The use of features from fluorescence, thermography, and NDVI imaging to detect biotic stress in lettuce. Plant Dis. 2018, 102, 1101–1107. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Chen, W.; Sankaran, S. High-throughput field phenotyping of Ascochyta blight disease severity in chickpea. Crop Prot. 2019, 125, 104885. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, C.; Guo, W.; Zhang, L.; Zhang, D. Automatic estimation of crop disease severity levels based on vegetation index normalization. Remote Sens. 2020, 12, 1930. [Google Scholar] [CrossRef]
- Liu, L.; Dong, Y.; Huang, W.; Du, X.; Ma, H. Monitoring wheat Fusarium head blight using unmanned aerial vehicle hyperspectral imagery. Remote Sens. 2020, 12, 3811. [Google Scholar] [CrossRef]
- Hernández-Clemente, R.; Navarro-Cerrillo, R.M.; Suárez, L.; Morales, F.; Zarco-Tejada, P.J. Assessing structural effects on PRI for stress detection in conifer forests. Remote Sens. Environ. 2011, 115, 2360–2375. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Alisaac, E.; Al Masri, A.; Behmann, J.; Dehne, H.W.; Oerke, E.C. Comparison and combination of thermal, fluorescence, and hyperspectral imaging for monitoring Fusarium head blight of wheat on spikelet scale. Sensors 2019, 19, 2281. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, M.; Ibrahim, A.M.; Xue, Q.; Jung, J.; Chang, A.; Rudd, J.C.; Landivar, J. Assessing winter wheat foliage disease severity using aerial imagery acquired from small unmanned aerial vehicle (UAV). Comput. Electron. Agric. 2020, 176, 105665. [Google Scholar] [CrossRef]
- He, L.; Qi, S.L.; Duan, J.Z.; Guo, T.C.; Feng, W.; He, D.X. Monitoring of wheat powdery mildew disease severity using multiangle hyperspectral remote sensing. Trans. Geosci. Remote Sens. 2020, 59, 979–990. [Google Scholar] [CrossRef]
- Pane, C.; Manganiello, G.; Nicastro, N.; Cardi, T.; Carotenuto, F. Powdery mildew caused by Erysiphe cruciferarum on wild rocket (Diplotaxis tenuifolia): Hyperspectral imaging and machine learning modeling for non-destructive disease detection. Agriculture 2021, 11, 337. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Gholz, H.L. Exploring the relationship between reflectance red edge and chlorophyll content in slash pine. Tree Physiol. 1990, 7, 33–48. [Google Scholar] [CrossRef]
- Liu, J.; van Iersel, M.W. Photosynthetic physiology of blue, green, and red light: Light intensity effects and underlying mechanisms. Front. Plant Sci. 2021, 12, 619987. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Gitelson, A.; Lang, M. Non-destructive determination of chlorophyll content of leaves of a green and an aurea mutant of tobacco by reflectance measurements. J. Plant Physiol. 1996, 148, 483–493. [Google Scholar] [CrossRef]
- Heber, U.; Shuvalov, V.A. Photochemical reactions of chlorophyll in dehydrated Photosystem II: Two chlorophyll forms (680 and 700 nm). Photosynth. Res. 2005, 84, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Guo, J.; Cui, Y.; Lü, T.; Zhang, X.; Shi, G. Effects of cadmium and salicylic acid on growth, spectral reflectance and photosynthesis of castor bean seedlings. Plant Soil 2011, 344, 131–141. [Google Scholar] [CrossRef]
- Iqbal, N.; Czékus, Z.; Ördög, A.; Poór, P. Ethylene-dependent effects of fusaric acid on the photosynthetic activity of tomato plants. Photosynthetica 2021, 59, 337–348. [Google Scholar] [CrossRef]
- Giraldo-Betancourt, C.; Velandia-Sánchez, E.A.; Fischer, G.; Gómez-Caro, S.; Martínez, L.J. Hyperspectral response of cape gooseberry (Physalis peruviana L.) plants inoculated with Fusarium oxysporum f. sp. physali for vascular wilt detection. Rev. Colomb. Cienc. Hortíc. 2020, 14, 301–313. [Google Scholar] [CrossRef]
- Ye, H.; Huang, W.; Huang, S.; Cui, B.; Dong, Y.; Guo, A.; Ren, Y.; Jin, Y. Recognition of Banana Fusarium Wilt based on UAV remote sensing. Remote Sens. 2020, 12, 938. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Liu, X.; Wu, W. A Hyperspectral Bidirectional Reflectance Model for Land Surface. Sensors 2020, 20, 4456. [Google Scholar] [CrossRef]











| DSC 1 vs. DSC 2 | |||||
| Df | Sum of Sqs | R2 | F | Pr (>F) | |
| DSC | 1 | 159.8 | 0.02076 | 98.91 | 0.001 |
| dpi | 2 | 69.7 | 0.00905 | 21.57 | 0.001 |
| POTS | 13 | 2170.9 | 0.2819 | 103.33 | 0.001 |
| Residual | 3280 | 5300.6 | 0.6883 | ||
| Total | 3296 | 7701.1 | 1 | ||
| DSC 1 vs. DSC 3 | |||||
| Df | Sum of Sqs | R2 | F | Pr (>F) | |
| DSC | 1 | 6054.3 | 0.43793 | 4308.88 | 0.001 |
| dpi | 2 | 584.1 | 0.04225 | 207.85 | 0.001 |
| POTS | 15 | 2041 | 0.14763 | 96.84 | 0.001 |
| Residual | 3662 | 5145.4 | 0.37219 | ||
| Total | 3680 | 13824.8 | 1 | ||
| DSC 2 vs. DSC 3 | |||||
| Df | Sum of Sqs | R2 | F | Pr (>F) | |
| DSC | 1 | 2958.5 | 0.28265 | 1331.24 | 0.001 |
| dpi | 1 | 343.8 | 0.03285 | 154.72 | 0.001 |
| POTS | 10 | 1348.8 | 0.12886 | 60.69 | 0.001 |
| Residual | 2617 | 5816 | 0.55564 | ||
| Total | 2629 | 10467.3 | 1 | ||
| Model with Correction for Class Imbalance | Model without Correction for Class Imbalance | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Observations | Observations | ||||||||||
| Predictions | d0 | d1 | d2 | d3 | Predictions | d0 | d1 | d2 | d3 | ||
| d0 | 6864 | 276 | 43 | 14 | d0 | 7620 | 416 | 122 | 14 | ||
| d1 | 673 | 157 | 81 | 0 | d1 | 68 | 20 | 47 | 7 | ||
| d2 | 158 | 105 | 228 | 182 | d2 | 7 | 102 | 181 | 202 | ||
| d3 | 0 | 0 | 9 | 756 | d3 | 0 | 0 | 11 | 729 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pane, C.; Manganiello, G.; Nicastro, N.; Carotenuto, F. Early Detection of Wild Rocket Tracheofusariosis Using Hyperspectral Image-Based Machine Learning. Remote Sens. 2022, 14, 84. https://doi.org/10.3390/rs14010084
Pane C, Manganiello G, Nicastro N, Carotenuto F. Early Detection of Wild Rocket Tracheofusariosis Using Hyperspectral Image-Based Machine Learning. Remote Sensing. 2022; 14(1):84. https://doi.org/10.3390/rs14010084
Chicago/Turabian StylePane, Catello, Gelsomina Manganiello, Nicola Nicastro, and Francesco Carotenuto. 2022. "Early Detection of Wild Rocket Tracheofusariosis Using Hyperspectral Image-Based Machine Learning" Remote Sensing 14, no. 1: 84. https://doi.org/10.3390/rs14010084