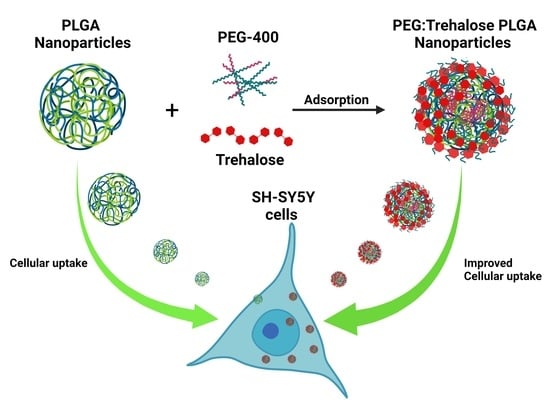
PEG 400:Trehalose Coating Enhances Curcumin-Loaded PLGA Nanoparticle Internalization in Neuronal Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation and Optimization of PLGA Nanoparticles
2.2.1. Nanoprecipitation Technique
2.2.2. Experimental Design
2.3. Curcumin Loading in Nanoparticles
2.4. Nanoparticles PEG:Trehalose Coating
2.5. Nanoparticles Characterization
2.5.1. Size, Polydispersity Index (Pdi), and Zeta Potential Measurements
2.5.2. Fourier-Transform Infrared Spectroscopy (FTIR)
2.5.3. Differential Scanning Calorimetry (DSC)
2.5.4. Thermogravimetric Analysis (TGA)
2.6. In Vitro Release Study of Curcumin
2.7. In Vitro Cytotoxicity Assay and Cellular Uptake
2.7.1. Cell Culture
2.7.2. In Vitro Cytotoxicity Assay
2.7.3. In Vitro Nanoparticles Cell Uptake
3. Results and Discussion
3.1. Optimization of Nanoprecipitation Technique for BNPs
3.2. Entrapment Efficiency
3.3. PEG:Trehalose Coating of Nanoparticles
3.3.1. Effect of PEG:Trehalose Coating in Blank Nanoparticles and Curcumin-Loaded Nanoparticles
3.3.2. Fourier Transform Infrared Spectra
3.3.3. Differential Scanning Calorimetry
3.3.4. Thermogravimetric Analysis
3.4. In Vitro Curcumin Release
3.5. In Vitro Cytotoxicity Assay
3.6. Nanoparticles Cell Uptake
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Del Prado-Audelo, M.L.; Caballero-Florán, I.H.; Meza-Toledo, J.A.; Mendoza-Muñoz, N.; González-Torres, M.; Florán, B.; Cortés, H.; Leyva-Gómez, G. Formulations of curcumin nanoparticles for brain diseases. Biomolecules 2019, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Cortés, H.; Reyes-Hernández, O.D.; Gonzalez-Torres, M.; Vizcaino-Dorado, P.A.; Del Prado-Audelo, M.L.; Alcalá-Alcalá, S.; Sharifi-Rad, J.; Figueroa-González, G.; Carmen, M.G.-D.; Florán, B.; et al. Curcumin for Parkinson’s disease: Potential therapeutic effects, molecular mechanisms, and nanoformulations to enhance its efficacy. Cell. Mol. Biol. 2021, 67, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Benameur, T.; Giacomucci, G.; Panaro, M.A.; Ruggiero, M.; Trotta, T.; Monda, V.; Pizzolorusso, I.; Lofrumento, D.D.; Porro, C.; Messina, G. New Promising Therapeutic Avenues of Curcumin in Brain Diseases. Molecules 2022, 27, 236. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, C.; Pan, H.; Li, L.; Yu, Y.; Liu, B. An insight into the in vivo imaging potential of curcumin analogues as fluorescence probes. Asian J. Pharm. Sci. 2021, 16, 419–431. [Google Scholar] [CrossRef]
- Nelson, K.M.; Dahlin, J.L.; Bisson, J.; Graham, J.; Pauli, G.F.; Walters, M.A. The Essential Medicinal Chemistry of Curcumin. J. Med. Chem. 2017, 60, 1620–1637. [Google Scholar] [CrossRef]
- Fonseca-Santos, B.; Gremião, M.P.D.; Chorilli, M. Nanotechnology-based drug delivery systems for the treatment of Alzheimer’s disease. Int. J. Nanomed. 2015, 10, 4981–5003. [Google Scholar] [CrossRef]
- Jamali, Z.; Khoobi, M.; Hejazi, S.M.; Eivazi, N.; Abdolahpour, S.; Imanparast, F.; Moradi-Sardareh, H.; Paknejad, M. Evaluation of targeted curcumin (CUR) loaded PLGA nanoparticles for in vitro photodynamic therapy on human glioblastoma cell line. Photodiagnosis Photodyn. Ther. 2018, 23, 190–201. [Google Scholar] [CrossRef]
- Swider, E.; Koshkina, O.; Tel, J.; Cruz, L.J.; de Vries, I.J.M.; Srinivas, M. Customizing poly(lactic-co-glycolic acid) particles for biomedical applications. Acta Biomater. 2018, 73, 38–51. [Google Scholar] [CrossRef]
- Essa, D.; Kondiah, P.P.D.; Choonara, Y.E.; Pillay, V. The Design of Poly(lactide-co-glycolide) Nanocarriers for Medical Applications. Front. Bioeng. Biotechnol. 2020, 8, 48. [Google Scholar] [CrossRef]
- Zhang, S.; Gao, H.; Bao, G. Physical Principles of Nanoparticle Cellular Endocytosis. ACS Nano 2015, 9, 8655–8671. [Google Scholar] [CrossRef]
- El-Hammadi, M.M.; Arias, J.L. Recent Advances in the Surface Functionalization of PLGA-Based Nanomedicines. Nanomaterials 2022, 12, 354. [Google Scholar] [CrossRef]
- Cheraga, N.; Sun, N.C.; Huang, X.X.; Ye, Z.; Xiao, Q.R.; Huang, N.P. Optimized rapamycin-loaded PEGylated PLGA nanoparticles: Preparation, characterization and pharmacokinetics study. J. Drug Deliv. Sci. Technol. 2021, 61, 102144. [Google Scholar] [CrossRef]
- Zhou, Y.Z.; Alany, R.G.; Chuang, V.; Wen, J. Optimization of PLGA nanoparticles formulation containing L-DOPA by applying the central composite design. Drug Dev. Ind. Pharm. 2013, 39, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Chiesa, E.; Dorati, R.; Modena, T.; Conti, B.; Genta, I. Multivariate analysis for the optimization of microfluidics-assisted nanoprecipitation method intended for the loading of small hydrophilic drugs into PLGA nanoparticles. Int. J. Pharm. 2018, 536, 165–177. [Google Scholar] [CrossRef]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef] [PubMed]
- Cu, Y.; Saltzman, W.M. Controlled Surface Modification with Poly(ethylene)glycol Enhances Diffusion of PLGA Nanoparticles in Human Cervical Mucus. Mol. Pharm. 2009, 6, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Zhang, J.; Zhao, M.; Tang, S.; Cheng, X.; Zhang, W.; Li, W.; Liu, X.; Peng, H.; Wang, Q. Effects of polyethylene glycol on the surface of nanoparticles for targeted drug delivery. Nanoscale 2021, 13, 10748–10764. [Google Scholar] [CrossRef]
- Vallorz, E.L.; Encinas-Basurto, D.; Schnellmann, R.G.; Mansour, H.M. Design, Development, Physicochemical Characterization, and In Vitro Drug Release of Formoterol PEGylated PLGA Polymeric Nanoparticles. Pharmaceutics 2022, 14, 638. [Google Scholar] [CrossRef]
- Mehanny, M.; Hathout, R.M.; Geneidi, A.S.; Mansour, S. Studying the effect of physically-adsorbed coating polymers on the cytotoxic activity of optimized bisdemethoxycurcumin loaded-PLGA nanoparticles. J. Biomed. Mater. Res. Part A 2017, 105, 1433–1445. [Google Scholar] [CrossRef]
- Mistry, A.; Stolnik, S.; Illum, L. Nanoparticles for direct nose-to-brain delivery of drugs. Int. J. Pharm. 2009, 379, 146–157. [Google Scholar] [CrossRef]
- Wang, Y.-Y.; Lai, S.K.; Suk, J.S.; Pace, A.; Cone, R.; Hanes, J. Addressing the PEG Mucoadhesivity Paradox to Engineer Nanoparticles that “Slip” through the Human Mucus Barrier. Angew. Chem. Int. Ed. 2008, 47, 9726–9729. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.K.; O’Hanlon, D.E.; Harrold, S.; Man, S.T.; Wang, Y.Y.; Cone, R.; Hanes, J. Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus. Proc. Natl. Acad. Sci. USA 2007, 104, 1482–1487. [Google Scholar] [CrossRef] [PubMed]
- Yoshizane, C.; Mizote, A.; Yamada, M.; Arai, N.; Arai, S.; Maruta, K.; Mitsuzumi, H.; Ariyasu, T.; Ushio, S.; Fukuda, S. Glycemic, insulinemic and incretin responses after oral trehalose ingestion in healthy subjects. Nutr. J. 2017, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.; Debnath, K.; Jana, N.R.; Jana, N.R. Trehalose-Functionalized Gold Nanoparticle for Inhibiting Intracellular Protein Aggregation. Langmuir 2017, 33, 13996–14003. [Google Scholar] [CrossRef] [PubMed]
- Siddhanta, S.; Zheng, C.; Narayana, C.; Barman, I. An impediment to random walk: Trehalose microenvironment drives preferential endocytic uptake of plasmonic nanoparticles. Chem. Sci. 2016, 7, 3730–3736. [Google Scholar] [CrossRef]
- Tosi, G.; Costantino, L.; Rivasi, F.; Ruozi, B.; Leo, E.; Vergoni, A.V.; Tacchi, R.; Bertolini, A.; Vandelli, M.A.; Forni, F. Targeting the central nervous system: In vivo experiments with peptide-derivatized nanoparticles loaded with Loperamide and Rhodamine-123. J. Control. Release 2007, 122, 1–9. [Google Scholar] [CrossRef]
- Montgomery, D.C. Design and Analysis of Experiments, 5th ed.; John Wiley: New York, NY, USA, 2001; ISBN 0471316490; 9780471316497. [Google Scholar]
- Ossama, M.; Hathout, R.M.; Attia, D.A.; Mortada, N.D. Augmented cytotoxicity using the physical adsorption of Poloxamer 188 on allicin-loaded gelatin nanoparticles. J. Pharm. Pharmacol. 2021, 73, 664–672. [Google Scholar] [CrossRef]
- Zhang, Y.; Huo, M.; Zhou, J.; Zou, A.; Li, W.; Yao, C.; Xie, S. DDSolver: An Add-In Program for Modeling and Comparison of Drug Dissolution Profiles. AAPS J. 2010, 12, 263–271. [Google Scholar] [CrossRef]
- Xicoy, H.; Wieringa, B.; Martens, G.J.M. The SH-SY5Y cell line in Parkinson’s disease research: A systematic review. Mol. Neurodegener. 2017, 12, 10. [Google Scholar] [CrossRef]
- Escobar, S.J.D.M.; Simone, M.; Martin, N.; Ribeiro, C.A.D.O.; Martinez, G.R.; Winnischofer, S.M.B.; Witting, P.K.; Rocha, M.E.M. Cytotoxic effects of 4′-hydroxychalcone on human neuroblastoma cells (SH-SY5Y). Toxicol. Vitr. 2019, 61, 104640. [Google Scholar] [CrossRef]
- García-Melero, J.; López-Mitjavila, J.-J.; García-Celma, M.J.; Rodriguez-Abreu, C.; Grijalvo, S. Rosmarinic Acid-Loaded Polymeric Nanoparticles Prepared by Low-Energy Nano-Emulsion Templating: Formulation, Biophysical Characterization, and In Vitro Studies. Materials 2022, 15, 4572. [Google Scholar] [CrossRef] [PubMed]
- Bacanlı, M.; Eşïm, Ö.; Erdoğan, H.; Sarper, M.; Erdem, O.; Özkan, Y. Evaluation of cytotoxic and genotoxic effects of paclitaxel-loaded PLGA nanoparticles in neuroblastoma cells. Food Chem. Toxicol. 2021, 154, 112323. [Google Scholar] [CrossRef] [PubMed]
- Lantto, T.A.; Colucci, M.; Závadová, V.; Hiltunen, R.; Raasmaja, A. Cytotoxicity of curcumin, resveratrol and plant extracts from basil, juniper, laurel and parsley in SH-SY5Y and CV1-P cells. Food Chem. 2009, 117, 405–411. [Google Scholar] [CrossRef]
- Teleanu, D.M.; Negut, I.; Grumezescu, V.; Grumezescu, A.M.; Teleanu, R.I. Nanomaterials for drug delivery to the central nervous system. Nanomaterials 2019, 9, 371. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, H.; El Kurdi, R.; Patra, D. Curcumin-PLGA based nanocapsule for the fluorescence spectroscopic detection of dopamine. RSC Adv. 2022, 12, 28245–28253. [Google Scholar] [CrossRef]
- Jahromi, L.P.; Ghazali, M.; Ashrafi, H.; Azadi, A. A comparison of models for the analysis of the kinetics of drug release from PLGA-based nanoparticles. Heliyon 2020, 6, e03451. [Google Scholar] [CrossRef]
- Vakilinezhad, M.A.; Amini, A.; Dara, T.; Alipour, S. Methotrexate and Curcumin co-encapsulated PLGA nanoparticles as a potential breast cancer therapeutic system: In vitro and in vivo evaluation. Colloids Surf. B Biointerfaces 2019, 184, 110515. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, M.; Wang, M.; Peng, H.; Hua, Q.; Ma, L.; Wang, B.; Wei, H. Facile Fabrication of 10-Hydroxycamptothecin-Backboned Amphiphilic Polyprodrug with Precisely Tailored Drug Loading Content for Controlled Release. Bioconjug. Chem. 2018, 29, 2239–2247. [Google Scholar] [CrossRef]
- Nair, K.L.; Thulasidasan, A.K.T.; Deepa, G.; Anto, R.J.; Kumar, G.S.V. Purely aqueous PLGA nanoparticulate formulations of curcumin exhibit enhanced anticancer activity with dependence on the combination of the carrier. Int. J. Pharm. 2012, 425, 44–52. [Google Scholar] [CrossRef]
- Tsai, Y.M.; Chien, C.F.; Lin, L.C.; Tsai, T.H. Curcumin and its nano-formulation: The kinetics of tissue distribution and blood-brain barrier penetration. Int. J. Pharm. 2011, 416, 331–338. [Google Scholar] [CrossRef]
- Chereddy, K.K.; Coco, R.; Memvanga, P.B.; Ucakar, B.; Des Rieux, A.; Vandermeulen, G.; Préat, V. Combined effect of PLGA and curcumin on wound healing activity. J. Control. Release 2013, 171, 208–215. [Google Scholar] [CrossRef] [PubMed]
- Ohtake, S.; Wang, Y.J. Trehalose: Current use and future applications. J. Pharm. Sci. 2011, 100, 2020–2053. [Google Scholar] [CrossRef] [PubMed]
- Khalifeh, M.; Barreto, G.E.; Sahebkar, A. Trehalose as a promising therapeutic candidate for the treatment of Parkinson’s disease. Br. J. Pharmacol. 2019, 176, 1173–1189. [Google Scholar] [CrossRef] [PubMed]
- Fule, R.; Kaleem, M.; Asar, T.O.; Rashid, M.A.; Shaik, R.A.; Eid, B.G.; Nasrullah, M.Z.; Ahmad, A.; Kazmi, I. Formulation, Optimization and Evaluation of Cytarabine-Loaded Iron Oxide Nanoparticles: From In Vitro to In Vivo Evaluation of Anticancer Activity. Nanomaterials 2023, 13, 175. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.I.; Samad, N.A.; Fang, L.; Lim, V. Cytotoxicity of targeted PLGA nanoparticles: A systematic review. RSC Adv. 2021, 11, 9433–9449. [Google Scholar] [CrossRef]
- Silva, A.T.C.R.; Cardoso, B.C.O.; e Silva, M.E.S.R.; Freitas, R.F.S.; Sousa, R.G. Characterization, and Study of PLGA Copolymer in Vitro Degradation. J. Biomater. Nanobiotechnol. 2015, 06, 8–19. [Google Scholar] [CrossRef]
- Mohan, P.R.K.; Sreelakshmi, G.; Muraleedharan, C.V.; Joseph, R. Water soluble complexes of curcumin with cyclodextrins: Characterization by FT-Raman spectroscopy. Vib. Spectrosc. 2012, 62, 77–84. [Google Scholar] [CrossRef]
- Ismail, E.H.; Sabry, D.Y.; Mahdy, H.; Khalil, M.M.H. Synthesis and Characterization of some Ternary Metal Complexes of Curcumin with 1,10-phenanthroline and their Anticancer Applications. J. Sci. Res. 2014, 6, 509–519. [Google Scholar] [CrossRef]
- Pietra, R.C.C.d.S.; Cruz, R.C.; Melo, C.N.; Rodrigues, L.B.; Santos, P.C.; Bretz, G.P.M.; Soares, B.M.; de Sousa, G.R.; Ferreira, M.V.L.; Cisalpino, P.S.; et al. Evaluation of polymeric PLGA nanoparticles conjugated to curcumin for use in aPDT. Braz. J. Pharm. Sci. 2017, 53, 1–9. [Google Scholar] [CrossRef]
- Jiang, G.; Jia, H.; Qiu, J.; Mo, Z.; Wen, Y.; Zhang, Y.; Wen, Y.; Xie, Q.; Ban, J.; Lu, Z.; et al. Plga nanoparticle platform for trans-ocular barrier to enhance drug delivery: A comparative study based on the application of oligosaccharides in the outer membrane of carriers. Int. J. Nanomed. 2020, 15, 9373–9387. [Google Scholar] [CrossRef]
- Akao, K.I.; Okubo, Y.; Asakawa, N.; Inoue, Y.; Sakurai, M. Infrared spectroscopic study on the properties of the anhydrous form II of trehalose. Implications for the functional mechanism of trehalose as a biostabilizer. Carbohydr. Res. 2001, 334, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Mu, L.; Wu, J.; Matsakas, L.; Chen, M.; Vahidi, A.; Grahn, M.; Rova, U.; Christakopoulos, P.; Zhu, J.; Shi, Y. Lignin from hardwood and softwood biomass as a lubricating additive to ethylene glycol. Molecules 2018, 23, 537. [Google Scholar] [CrossRef] [PubMed]
- Marcos, M.; Cabaleiro, D.; Guimarey, M.; Comuñas, M.; Fedele, L.; Fernández, J.; Lugo, L. PEG 400-Based Phase Change Materials Nano-Enhanced with Functionalized Graphene Nanoplatelets. Nanomaterials 2017, 8, 16. [Google Scholar] [CrossRef] [PubMed]
- Cherecheş, M.; Bejan, D.; Cherecheş, E.I.; Minea, A.A. Experimental studies on several properties of PEG 400 and MWCNT nano-enhanced PEG 400 fluids. J. Mol. Liq. 2022, 356, 119049. [Google Scholar] [CrossRef]
- Yang, J.M.; Su, W.Y.; Leu, T.L.; Yang, M.C. Evaluation of chitosan/PVA blended hydrogel membranes. J. Membr. Sci. 2004, 236, 39–51. [Google Scholar] [CrossRef]
- Fugita, R.A.; Gálico, D.A.; Guerra, R.B.; Perpétuo, G.L.; Treu-Filho, O.; Galhiane, M.S.; Mendes, R.A.; Bannach, G. Thermal behaviour of curcumin. Braz. J. Therm. Anal. 2012, 1, 19–23. [Google Scholar]
- Verhoeven, N.; Neoh, T.L.; Furuta, T.; Yamamoto, C.; Ohashi, T.; Yoshii, H. Characteristics of dehydration kinetics of dihydrate trehalose to its anhydrous form in ethanol by DSC. Food Chem. 2012, 132, 1638–1643. [Google Scholar] [CrossRef]
- Akl, M.A.; Kartal-Hodzic, A.; Oksanen, T.; Ismael, H.R.; Afouna, M.M.; Yliperttula, M.; Samy, A.M.; Viitala, T. Factorial design formulation optimization and in vitro characterization of curcumin-loaded PLGA nanoparticles for colon delivery. J. Drug Deliv. Sci. Technol. 2016, 32, 10–20. [Google Scholar] [CrossRef]
- Sanna, V.; Roggio, A.M.; Posadino, A.M.; Cossu, A.; Marceddu, S.; Mariani, A.; Alzari, V.; Uzzau, S.; Pintus, G.; Sechi, M. Novel docetaxel-loaded nanoparticles based on poly(lactide-co-caprolactone) and poly(lactide-co-glycolide-co-caprolactone) for prostate cancer treatment: Formulation, characterization, and cytotoxicity studies. Nanoscale Res. Lett. 2011, 6, 260. [Google Scholar] [CrossRef]
- Putro, P.A.; Sulaeman, A.S.; Maddu, A. Polyvinyl alcohol-based hydrogel: A systematic literature review on thermal properties by differential scanning calorimetry. J. Phys. Conf. Ser. 2021, 2019, 012101. [Google Scholar] [CrossRef]
- Del Prado-Audelo, M.L.; Magaña, J.J.; Mejía-Contreras, B.A.; Borbolla-Jiménez, F.V.; Giraldo-Gomez, D.M.; Piña-Barba, M.C.; Quintanar-Guerrero, D.; Leyva-Gómez, G. In vitro cell uptake evaluation of curcumin-loaded PCL/F68 nanoparticles for potential application in neuronal diseases. J. Drug Deliv. Sci. Technol. 2019, 52, 905–914. [Google Scholar] [CrossRef]
- Kaur, A.; Rathee, J.; Kanwar, R.; Kaushik, D.; Salunke, D.B.; Mehta, S.K. TLR2 agonistic lipopeptide enriched PLGA nanoparticles as combinatorial drug delivery vehicle. Colloids Surf. A Physicochem. Eng. Asp. 2022, 647, 129084. [Google Scholar] [CrossRef]
- Gomaa, M.M.; Hugenschmidt, C.; Dickmann, M.; Abdel-Hady, E.E.; Mohamed, H.F.M.; Abdel-Hamed, M.O. Crosslinked PVA/SSA proton exchange membranes: Correlation between physiochemical properties and free volume determined by positron annihilation spectroscopy. Phys. Chem. Chem. Phys. 2018, 20, 28287–28299. [Google Scholar] [CrossRef] [PubMed]
- Palacios, J.; Albano, C.; González, G.; Castillo, R.V.; Karam, A.; Covis, M. Characterization and Thermal Degradation of Poly(d,l-Lactide-co-Glycolide) Composites with Nanofillers. Polym. Eng. Sci. 2013, 53, 1414–1429. [Google Scholar] [CrossRef]
- Masek, A.; Chrzescijanska, E.; Zaborski, M. Characteristics of curcumin using cyclic voltammetry, UV-vis, fluorescence and thermogravimetric analysis. Electrochim. Acta 2013, 107, 441–447. [Google Scholar] [CrossRef]
- Rathee, J.; Kaur, A.; Kanwar, R.; Kaushik, D.; Kumar, R.; Salunke, D.B.; Mehta, S.K. Polymeric Nanoparticles as a Promising Drug Delivery Platform for the Efficacious Delivery of Toll-Like Receptor 7/8 Agonist and IDO-Inhibitor. Colloids Surf. A Physicochem. Eng. Asp. 2022, 632, 127764. [Google Scholar] [CrossRef]
- Albisa, A.; Piacentini, E.; Sebastian, V.; Arruebo, M.; Santamaria, J.; Giorno, L. Preparation of Drug-Loaded PLGA-PEG Nanoparticles by Membrane-Assisted Nanoprecipitation. Pharm. Res. 2017, 34, 1296–1308. [Google Scholar] [CrossRef]
- Costa, P.; Lobo, J.M.S. Modeling and comparison of dissolution profiles. Eur. J. Pharm. Sci. 2001, 13, 123–133. [Google Scholar] [CrossRef]
- Kakran, M.; Sahoo, N.G.; Li, L.; Judeh, Z. Fabrication of quercetin nanoparticles by anti-solvent precipitation method for enhanced dissolution. Powder Technol. 2012, 223, 59–64. [Google Scholar] [CrossRef]
- Heredia, N.S.; Vizuete, K.; Flores-Calero, M.; Pazmiño, V.K.; Pilaquinga, F.; Kumar, B.; Debut, A. Comparative statistical analysis of the release kinetics models for nanoprecipitated drug delivery systems based on poly(lactic-co-glycolic acid). PLoS ONE 2022, 17, e0264825. [Google Scholar] [CrossRef]
- Peppas, N.A.; Sahlin, J.J. A simple equation for the description of solute release. III. Coupling of diffusion and relaxation. Int. J. Pharm. 1989, 57, 169–172. [Google Scholar] [CrossRef]
- Siepmann, J.; Peppas, N.A. Modeling of drug release from delivery systems based on hydroxypropyl methylcellulose (HPMC). Adv. Drug Deliv. Rev. 2001, 48, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Mathematical models of drug release. In Strategies to Modify the Drug Release from Pharmaceutical Systems; Elsevier: Amsterdam, The Netherlands, 2015; pp. 63–86. ISBN 9780081000922.
- Han, F.Y.; Liu, Y.; Kumar, V.; Xu, W.; Yang, G.; Zhao, C.X.; Woodruff, T.M.; Whittaker, A.K.; Smith, M.T. Sustained-release ketamine-loaded nanoparticles fabricated by sequential nanoprecipitation. Int. J. Pharm. 2020, 581, 119291. [Google Scholar] [CrossRef] [PubMed]
- Namkaew, J.; Jaroonwitchawan, T.; Rujanapun, N.; Saelee, J.; Noisa, P. Combined effects of curcumin and doxorubicin on cell death and cell migration of SH-SY5Y human neuroblastoma cells. Vitr. Cell. Dev. Biol. Anim. 2018, 54, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.S.; Boddapati, S.; Emadi, S.; Sierks, M.R. Curcumin reduces α-synuclein induced cytotoxicity in Parkinson’s disease cell model. BMC Neurosci. 2010, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Sartaj, A.; Qamar, Z.; Qizilbash, F.F.; Annu; Shadab; Alhakamy, N.A.; Baboota, S.; Ali, J. Polymeric nanoparticles: Exploring the current drug development and therapeutic insight of breast cancer treatment and recommendations. Polymers 2021, 13, 4400. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Wu, J.; Li, H.; Ma, G. Recent research and development of PLGA/PLA microspheres/nanoparticles: A review in scientific and industrial aspects. Front. Chem. Sci. Eng. 2019, 13, 14–27. [Google Scholar] [CrossRef]
- Doggui, S.; Sahni, J.K.; Arseneault, M.; Dao, L.; Ramassamy, C. Neuronal uptake and neuroprotective effect of curcumin-loaded PLGA nanoparticles on the human SK-N-SH cell line. J. Alzheimer. Dis. 2012, 30, 377–392. [Google Scholar] [CrossRef]
- Paka, G.D.; Ramassamy, C. Optimization of Curcumin-Loaded PEG-PLGA Nanoparticles by GSH Functionalization: Investigation of the Internalization Pathway in Neuronal Cells. Mol. Pharm. 2017, 14, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.N.; Ha, P.T.; Nguyen, A.S.; Nguyen, D.T.; Do, H.D.; Thi, Q.N.; Thi, M.N.H. Curcumin as fluorescent probe for directly monitoring in vitro uptake of curcumin combined paclitaxel loaded PLA-TPGS nanoparticles. Adv. Nat. Sci. Nanosci. Nanotechnol. 2016, 7, 025001. [Google Scholar] [CrossRef]
- Moustapha, A.; Pérétout, P.; Rainey, N.; Sureau, F.; Geze, M.; Petit, J.-M.; Dewailly, E.; Slomianny, C.; Petit, P. Curcumin induces crosstalk between autophagy and apoptosis mediated by calcium release from the endoplasmic reticulum, lysosomal destabilization and mitochondrial events. Cell Death Discov. 2015, 1, 15017. [Google Scholar] [CrossRef] [PubMed]
- Stewart, S.; He, X. Intracellular Delivery of Trehalose for Cell Banking. Langmuir 2019, 35, 7414–7422. [Google Scholar] [CrossRef] [PubMed]
- Cunha, A.; Gaubert, A.; Verget, J.; Thiolat, M.L.; Barthélémy, P.; Latxague, L.; Dehay, B. Trehalose-Based Nucleolipids as Nanocarriers for Autophagy Modulation: An In Vitro Study. Pharmaceutics 2022, 14, 857. [Google Scholar] [CrossRef]
- DeBosch, B.J.; Heitmeier, M.R.; Mayer, A.L.; Higgins, C.B.; Crowley, J.R.; Kraft, T.E.; Chi, M.; Newberry, E.P.; Chen, Z.; Finck, B.N.; et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci. Signal. 2016, 9, ra21. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.S.; Cho, E.D.; Ahn, W.J.; Lee, K.W.; Lee, S.J.; Lee, H.J. Is trehalose an autophagic inducer? Unraveling the roles of non-reducing disaccharides on autophagic flux and alpha-synuclein aggregation. Cell Death Dis. 2017, 8, e3091. [Google Scholar] [CrossRef] [PubMed]










| Codified Value | PLGA (mg) (x1) | PVA (%w/v) (x2) | Stirring (rpm) (x3) |
|---|---|---|---|
| −1.68 (−α) | 7.95 | 0.32 | 630 |
| −1 | 10.0 | 1.0 | 800 |
| 0 | 13.0 | 2.0 | 1050 |
| 1 | 16.0 | 3.0 | 1300 |
| +1.68 (+α) | 18.04 | 3.68 | 1470 |
| Independent Variables | Response | ||||
|---|---|---|---|---|---|
| Run | PLGA (mg) (x1) | PVA (%w/v) (x2) | Stirring (rpm) (x3) | Size (nm) (y1) | Pdi (y2) |
| 1 | 18.04 | 2 | 1050 | 227.78 ± 11.561 | 0.160 ± 0.019 |
| 2 | 10 | 3 | 1300 | 179.63 ± 12.903 | 0.222 ± 0.067 |
| 3 | 13 | 2 | 1050 | 213.38 ± 5.298 | 0.133 ± 0.033 |
| 4 | 7.95 | 2 | 1050 | 190.74 ± 12.377 | 0.249 ± 0.042 |
| 5 | 10 | 1 | 1300 | 174.33 ± 2.972 | 0.201 ± 0.024 |
| 6 | 13 | 2 | 1050 | 208.83 ± 5.266 | 0.183 ± 0.020 |
| 7 | 13 | 2 | 629.5 | 226.18 ± 5.360 | 0.120 ± 0.027 |
| 8 | 13 | 2 | 1050 | 215.06 ± 6.057 | 0.155 ± 0.026 |
| 9 | 13 | 0.3 | 1050 | 210.29 ± 2.701 | 0.163 ± 0.013 |
| 10 | 13 | 2 | 1470.4 | 184.56 ± 5.593 | 0.240 ± 0.023 |
| 11 | 16 | 1 | 1300 | 205.76 ± 2.225 | 0.196 ± 0.009 |
| 12 | 13 | 3.6 | 1050 | 216.63 ± 6.060 | 0.157 ± 0.028 |
| 13 | 10 | 1 | 800 | 186.61 ± 2.601 | 0.222 ± 0.074 |
| 14 | 16 | 1 | 800 | 217.96 ± 3.743 | 0.139 ± 0.007 |
| 15 | 13 | 2 | 1050 | 197.02 ± 2.605 | 0.164 ± 0.013 |
| 16 | 16 | 3 | 800 | 235.18 ± 5.824 | 0.159 ± 0.019 |
| 17 | 13 | 2 | 1050 | 194.69 ± 2.152 | 0.166 ± 0.011 |
| 18 | 13 | 2 | 1050 | 203.71 ± 1.438 | 0.163 ±0.013 |
| 19 | 10 | 3 | 800 | 185.62 ± 5.867 | 0.208 ± 0.043 |
| 20 | 16 | 3 | 1300 | 201.37 ± 6.279 | 0.174 ± 0.018 |
| Source | Sum of Squares | d.f. | Mean Square | F-Ratio | p-Value |
|---|---|---|---|---|---|
| x1: PLGA | 8468.26 | 1 | 8468.26 | 95.58 | 0.0000 |
| x2: PVA | 169.88 | 1 | 169.88 | 1.92 | 0.1864 |
| x3: SpeedStirring | 4806.13 | 1 | 4806.13 | 54.25 | 0.0000 |
| x1 x1 | 46.34 | 1 | 46.34 | 0.52 | 0.4807 |
| x1 x2 | 27.22 | 1 | 27.22 | 0.31 | 0.5875 |
| x1x3 | 74.55 | 1 | 74.55 | 0.84 | 0.3735 |
| x2 x2 | 8.72 | 1 | 8.72 | 0.10 | 0.7581 |
| x2 x3 | 314.65 | 1 | 314.65 | 3.55 | 0.0790 |
| x3x3 | 251.48 | 1 | 251.48 | 2.84 | 0.1127 |
| Blocks | 174.11 | 2 | 87.05 | 0.98 | 0.3972 |
| Lack of fit | 3154.95 | 33 | 95.60 | 1.08 | 0.4543 |
| Pure error | 1328.95 | 15 | 88.60 | ||
| Total | 18,823.30 | 59 |
| Source | Sum of Squares | d.f. | Mean Square | F-Ratio | p-Value |
|---|---|---|---|---|---|
| x1: PLGA | 0.024360 | 1 | 0.024360 | 30.45 | 0.0001 |
| x2: PVA | 0.000003 | 1 | 0.000003 | 0.00 | 0.9468 |
| x3: SpeedStirring | 0.005517 | 1 | 0.005517 | 6.90 | 0.0191 |
| x1 x1 | 0.012436 | 1 | 0.012436 | 15.55 | 0.0013 |
| x1 x2 | 0.000032 | 1 | 0.000032 | 0.04 | 0.8426 |
| x1x3 | 0.012880 | 1 | 0.012880 | 16.10 | 0.0011 |
| x2 x2 | 0.000078 | 1 | 0.000078 | 0.10 | 0.7588 |
| x2 x3 | 0.005046 | 1 | 0.005046 | 6.31 | 0.0240 |
| x3x3 | 0.003148 | 1 | 0.003148 | 3.94 | 0.0659 |
| Blocks | 0.003446 | 2 | 0.001723 | 2.15 | 0.1506 |
| Lack of fit | 0.054255 | 33 | 0.001644 | 2.06 | 0.0693 |
| Pure error | 0.011999 | 15 | 0.000799 | ||
| Total | 0.132069 | 59 |
| Particles | Parameters | Zero Order | First Order | Higuchi | Korsmeyer–Peppas | Hixon–Crowell |
|---|---|---|---|---|---|---|
| f = k × t | ln(1 − f) = −kt | f = kt1/2 | F = kKP·tn | 1 − 1(1 − f)1/3 = −k × t | ||
| CNPs | r2 | 0.756 | 0.833 | 0.953 | 0.967 | 0.812 |
| AIC | 93.5 | 86.2 | 70.2 | 68.7 | 88.6 | |
| MSC | 1.40 | 1.92 | 3.06 | 3.17 | 1.75 | |
| CNPs-PT | r2 | 0.598 | 0.689 | 0.895 | 0.953 | 0.656 |
| AIC | 96.4 | 93.6 | 79.3 | 71.2 | 94.39 | |
| MSC | 1.02 | 1.21 | 2.24 | 2.82 | 1.16 | |
| Particles | Parameters | Makoid–Banakar | Peppas–Shalin | Baker–Lonsdale | Weibull | |
| f = kMB t1/2e(−ct) | f = k1 × t1/2+k2·t | 3/2 ∗ [1 − (1 − F/100)^(2/3)] − F/100 = kBL ∗ t | f = Fmax·[1 − e(−tˆβα)] | |||
| CNPs | r2 | 0.969 | 0.969 | 0.952 | 0.968 | |
| AIC | 69.6 | 71.3 | 71.3 | 70.3 | ||
| MSC | 3.11 | 3.10 | 2.9 | 3.05 | ||
| CNPs-PT | r2 | 0.958 | 0.939 | 0.902 | 0.950 | |
| AIC | 71.9 | 75.0 | 78.3 | 73.9 | ||
| MSC | 2.76 | 2.54 | 2.31 | 2.63 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caballero-Florán, I.H.; Cortés, H.; Borbolla-Jiménez, F.V.; Florán-Hernández, C.D.; Del Prado-Audelo, M.L.; Magaña, J.J.; Florán, B.; Leyva-Gómez, G. PEG 400:Trehalose Coating Enhances Curcumin-Loaded PLGA Nanoparticle Internalization in Neuronal Cells. Pharmaceutics 2023, 15, 1594. https://doi.org/10.3390/pharmaceutics15061594
Caballero-Florán IH, Cortés H, Borbolla-Jiménez FV, Florán-Hernández CD, Del Prado-Audelo ML, Magaña JJ, Florán B, Leyva-Gómez G. PEG 400:Trehalose Coating Enhances Curcumin-Loaded PLGA Nanoparticle Internalization in Neuronal Cells. Pharmaceutics. 2023; 15(6):1594. https://doi.org/10.3390/pharmaceutics15061594
Chicago/Turabian StyleCaballero-Florán, Isaac H., Hernán Cortés, Fabiola V. Borbolla-Jiménez, Carla D. Florán-Hernández, María L. Del Prado-Audelo, Jonathan J. Magaña, Benjamín Florán, and Gerardo Leyva-Gómez. 2023. "PEG 400:Trehalose Coating Enhances Curcumin-Loaded PLGA Nanoparticle Internalization in Neuronal Cells" Pharmaceutics 15, no. 6: 1594. https://doi.org/10.3390/pharmaceutics15061594