HER2/neu Oncogene Silencing in a Breast Cancer Cell Model Using Cationic Lipid-Based Delivery Systems
Abstract
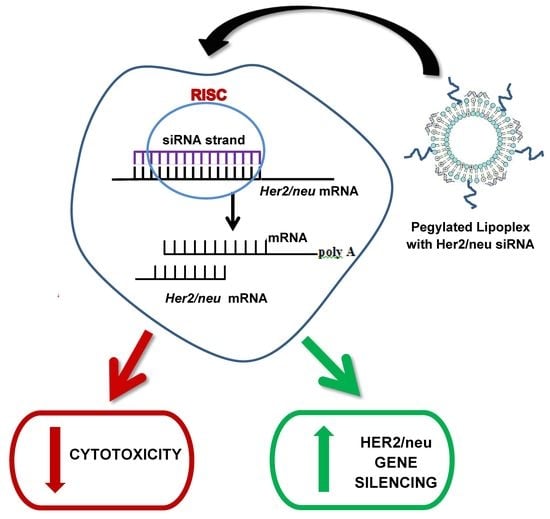
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Liposome Preparation
2.3. Liposome: siRNA Complex Preparation
2.4. Characterization
2.4.1. Transmission Electron Microscopy (TEM)
2.4.2. Size, Zeta Potential, and Polydispersity Index
2.5. Band-Shift Assay
2.6. Serum Nuclease Protection Assay
2.7. Cell Viability Studies
2.8. HER2/neu Silencing at mRNA and Protein Levels
2.8.1. siRNA Transfection
2.8.2. RNA Extraction and qRT-PCR
2.8.3. Protein Extraction and Western Blotting
2.9. Statistical Analysis
3. Results
3.1. Liposome and Lipoplex Characterization
3.2. Liposome Binding and Protection Efficiencies
3.3. Cell Viability Assay
3.4. HER2/neu Gene Silencing in SKBR-3 (Breast Cancer) Cells
3.4.1. Real-Time Quantitative PCR (qRT-PCR)
3.4.2. HER2/neu Protein Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer. (accessed on 10 January 2023).
- Rizvi, S.A.; Saleh, A.M. Applications of nanoparticle systems in drug delivery technology. Saudi Pharm. J. 2018, 26, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Menard, S.; Tagliabue, E.; Campiglio, M.; Pupa, S.M. Role of HER2 gene overexpression in breast carcinoma. J. Cell. Physiol. 2000, 182, 150–162. [Google Scholar] [CrossRef]
- Núñez, C.; Capelo, J.L.; Igrejas, G.; Alfonso, A.; Botana, L.M.; Lodeiro, C. An overview of the effective combination therapies for the treatment of breast cancer. Biomaterials 2016, 97, 34–50. [Google Scholar] [CrossRef]
- Padayachee, J.; Daniels, A.; Balgobind, A.; Ariatti, M.; Singh, M. HER2/neu and MYC Gene Silencing in breast cancer: Therapeutic potential and advancement in non-viral nanocarrier systems. Nanomedicine 2020, 15, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Tsé, C.; Gauchez, A.S.; Jacot, W.; Lamy, P.J. HER2 shedding and serum HER2 extracellular domain: Biology and clinical utility in breast cancer. Cancer Treat. Rev. 2012, 38, 133–142. [Google Scholar] [CrossRef]
- Yarden, Y. Biology of HER2 and its importance in breast cancer. Oncology 2001, 61, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Whitehead, K.A.; Langer, R.; Anderson, D.G. Knocking down barriers: Advances in siRNA delivery. Nat. Rev. Drug Discov. 2009, 8, 129–138. [Google Scholar] [CrossRef]
- Daniels, A.; Singh, M. Sterically stabilized siRNA: Gold nanocomplexes enhance c-MYC in a breast cancer cell model. Nanomedicine 2019, 14, 1387–1401. [Google Scholar] [CrossRef]
- Kesharwani, P.; Gajbhiye, V.; Jain, N.K. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials 2012, 33, 7138–7150. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.K.; Park, T.G. siRNA delivery systems for cancer treatment. Adv. Drug Deliv. Rev. 2009, 61, 850–862. [Google Scholar] [CrossRef]
- Shim, G.; Kim, M.G.; Park, J.Y.; Oh, Y.K. Application of cationic liposomes for delivery of nucleic acids. Asian, J. Pharm. Sci. 2013, 8, 72–80. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Li, S.; Li, Y.; Yang, G.; Li, M.; Xie, Y.; Su, W.; Wu, J.; Ma, W.; Li, H.; et al. Cationic liposomes co-deliver chemotherapeutics and siRNA for the treatment of breast cancer. Eur. J. Med. Chem. 2022, 233, 114198. [Google Scholar] [CrossRef]
- Vaidya, S.; Jeengar, M.K.; Wadaan, M.A.; Mahboob, S.; Kumar, P.; Reece, L.M.; Bathula, S.R.; Dutta, M. Design and In Vitro Evaluation of Novel Cationic Lipids for siRNA Delivery in Breast Cancer Cell Lines. Evid.-Based Complement. Altern. Med. 2022, 2022, 9231641. [Google Scholar] [CrossRef]
- Tenchov, R.; Bird, R.; Curtze, A.E.; Zhou, Q. Lipid Nanoparticles-From Liposomes to mRNA Vaccine Delivery, a Landscape of Research Diversity and Advancement. ACS Nano. 2021, 15, 16982–17015. [Google Scholar] [CrossRef]
- Gao, H.; Hui, K.M. Synthesis of a novel series of cationic lipids that can act as efficient gene delivery vehicles through systematic heterocyclic substitution of cholesterol derivatives. Gene Ther. 2001, 8, 855–863. [Google Scholar] [CrossRef] [Green Version]
- Elsana, H.; Olusanya, T.O.B.; Carr-wilkinson, J.; Darby, S.; Faheem, A.; Elkordy, A.A. Evaluation of novel cationic gene based liposomes with cyclodextrin prepared by thin film hydration and microfluidic systems. Sci. Rep. 2019, 9, 15120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hattori, Y.; Tang, M.; Torii, S.; Tomita, K.; Sagawa, A.; Inoue, N.; Yamagishi, R.; Ozaki, K.-I. Optimal combination of cationic lipid and phospholipid in cationic liposomes for gene knockdown in breast cancer cells and mouse lung using siRNA lipoplexes. Mol. Med. Rep. 2022, 26, 253. [Google Scholar] [CrossRef]
- Domínguez-Arca, V.; Sabín, J.; García-Río, L.; Bastos, M.; Taboada, P.; Barbosa, S.; Prieto, G. On the structure and stability of novel cationic DPPC liposomes doped with gemini surfactants. J. Mol. Liq. 2022, 366, 120230. [Google Scholar] [CrossRef]
- Corrie, L.; Gulati, M.; Awasthi, A.; Vishwas, S.; Kaur, J.; Khursheed, R.; Porwal, O.; Alam, A.; Parveen, S.R.; Singh, H.; et al. Harnessing the dual role of polysaccharides in treating gastrointestinal diseases: As therapeutics and polymers for drug delivery. Chem.-Bio.Interact. 2022, 368, 110238. [Google Scholar]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D.; et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Caracciolo, G. Liposome-protein corona in a physiological environment: Challenges and opportunities for targeted delivery of nanomedicines. Nanomed. Nanotechnol. Biol. Med. 2015, 11, 543–557. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaumet, M.; Vargas, A.; Gurny, R.; Delie, F. Nanoparticles for drug delivery: The need for precision in reporting particle size parameters. Eur. J. Pharm. Biopharm. 2008, 69, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.; Li, X. Liposomal delivery of CRISPR/Cas9. Cancer Gene Ther. 2020, 27, 515–527. [Google Scholar] [CrossRef]
- Singh, M.; Ariatti, M. Targeted gene delivery into HepG2 cells using complexes containing DNA, cationized asialoorosomucoid and activated cationic liposomes. J. Control. Release 2003, 92, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Ariatti, M. A cationic cytofectin with long spacer mediates favourable transfection in transformed human epithelial cells. Int. J. Pharm. 2006, 309, 189–198. [Google Scholar] [CrossRef]
- Gao, X.; Huang, L. A novel cationic liposome reagent for efficient transfection of mammalian cells. Biochem. Biophys. Res. Commun. 1991, 179, 280–285. [Google Scholar] [CrossRef]
- Singh, M. Assessing nucleic acid: Cationic nanoparticle interaction for gene delivery. In Bio-Carrier Vectors; Narayanan, K., Ed.; Springer: New York, NY, USA, 2021; Volume 2211, pp. 43–55. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kuntsche, J.; Horst, J.C.; Bunjes, H. Cryogenic transmission electron microscopy (cryo-TEM) for studying the morphology of colloidal drug delivery systems. Int. J. Pharm. 2011, 417, 120–137. [Google Scholar] [CrossRef]
- Lorenzer, C.; Dirin, M.; Winkler, A.M.; Baumann, V.; Winkler, J. Going beyond the liver: Progress and challenges of targeted delivery of siRNA therapeutics. J. Control. Release 2015, 203, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Resnier, P.; Montier, T.; Mathieu, V.; Benoit, J.P.; Passirani., C. A review of the current status of siRNA nanomedicines in the treatment of cancer. Biomaterials 2013, 34, 6429–6443. [Google Scholar] [CrossRef]
- Kapoor, M.; Burgess, D.J.; Patil, S.D. Physicochemical characterization techniques for lipid-based delivery systems for siRNA. Int. J. Pharm. 2012, 427, 35–57. [Google Scholar] [CrossRef]
- Mével, M.; Kamaly, N.; Carmona, S.; Oliver, M.H.; Jorgensen, M.R.; Crowther, C.; Salazar, F.H.; Marion, P.L.; Fujino, M.; Natori, Y.; et al. DODAG; a versatile new cationic lipid that mediates efficient delivery of pDNA and siRNA. J. Control. Release 2010, 143, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Kenworthy, A.K.; Hristova, K.; Needham, D.; McIntosh, T.J. Range and magnitude of the steric pressure between bilayers containing phospholipids with covalently attached poly(ethylene glycol). Biophys. J. 1995, 68, 1921–1936. [Google Scholar] [CrossRef] [Green Version]
- Needham, D.; McIntosh, T.J.; Lasic, D.D. Repulsive interactions and mechanical stability of polymer-grafted lipid membranes. Biochim. Biophys. Acta. 1992, 1108, 40–48. [Google Scholar] [CrossRef]
- Hassan, S.; Prakash, G.; Ozturk, A.B.; Saghazadeh, S.; Sohail, M.F.; Seo, J.; Dokmeci, M.R.; Zhang, Y.S.; Khademhosseini, A. Evolution and Clinical Translation of Drug Delivery Nanomaterials. Nano Today 2017, 15, 91–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, L.; Zhu, W.; Guo, R.; Sun, H.; Chen, X.; Deng, N. Barriers and Strategies of Cationic Liposomes for Cancer Gene Therapy. Mol. Ther.-Methods Clin. Dev. 2020, 18, 751–764. [Google Scholar] [PubMed]
- Danaei, M.; Dehghankhold, M.; Ataei, S.; Davarani, F.H.; Javanmard, R.; Dokhani, A.; Khorasani, S.; Mozafari, M.R. Impact of Particle Size and Polydispersity Index on the Clinical Applications of Lipidic Nanocarrier Systems. Pharmaceutics 2018, 10, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turetskiy, E.A.; Koloskova, O.O.; Nosova, A.S.; Shilovskiy, I.P.; Sebyakin, Y.L.; Khaitov, M.R. Physicochemical properties of lipopeptide-based liposomes and theircomplexes with siRNA. Biomed. Khim. 2017, 63, 472–475. [Google Scholar] [CrossRef] [Green Version]
- Belletti, D.; Tosi, G.; Forni, F.; Lagreca, I.; Barozzi, P.; Pederzoli, F.; Vandelli, M.A.; Riva, G.; Luppi, M.; Ruozi, B. PEGylated siRNA lipoplexes for silencing of BLIMP-1 in Primary Effusion Lymphoma: In vitro evidences of antitumoral activity. Eur. J. Pharm. Biopharm. 2016, 99, 7–17. [Google Scholar] [CrossRef] [Green Version]
- Pandi, P.; Jain, A.; Kommineni, N.; Ionov, M.; Bryszewska, M.; Khan, W. Dendrimer as a new potential carrier for topical delivery of siRNA: A comparative study of dendriplex vs. lipoplex for delivery of TNF-α siRNA. Int. J. Pharm. 2018, 550, 240–250. [Google Scholar] [CrossRef]
- Spagnou, S.; Miller, A.D.; Keller, M. Lipidic carriers of siRNA: Differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry 2004, 43, 13348–13356. [Google Scholar] [CrossRef] [PubMed]
- Van Gaal, E.V.B.; van Eijk, R.; Oosting, R.S.; Kok, R.J.; Hennink, W.E.; Crommelin, D.J.A.; Mastrobattista, E. How to screen non-viral gene delivery systems in vitro? J. Control. Release 2011, 154, 218–232. [Google Scholar] [CrossRef]
- Floch, V.; Loisel, S.; Guenin, E.; Herve, A.C.; Clement, J.C.; Yaouanc, J.J.; des Abbayes, H.; Ferec, C. Cation substitution in cationic phosphonolipids: A new concept to improve transfection activity and decrease cellular toxicity. J. Med. Chem. 2000, 30, 4617–4628. [Google Scholar] [CrossRef] [PubMed]
- Boomer, J.A.; Thompson, D.H.; Sullivan, S.M. Formation of plasmid based transfection complexes with an acid-labile cationic lipid: Characterization of in vitro and in vivo gene transfer. Pharm. Res. 2002, 19, 1292–1301. [Google Scholar] [CrossRef]
- Liu, D.L.; Hu, J.J.; Qiao, W.H.; Li, Z.S.; Zhang, S.B.; Cheng, L.B. Synthesis of carbamate-linked lipids for gene delivery. Bioorganic Med. Chem. Lett. 2005, 15, 3147–3150. [Google Scholar] [CrossRef]
- Monpara, J.; Velga, D.; Verma, T.; Gupta, S.; Vavia, P. Cationic cholesterol derivative efficiently delivers the genes: In silicoand in vitro studies. Drug Deliv. Transl. Res. 2019, 9, 106–122. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Sun, J.; Gao, J.; Liu, W.; Li, B.; Guo, Y.; Chen, J. DC-Chol/DOPE cationic liposomes: A comparative study of the influence factors on plasmid pDNA and siRNA gene delivery. Int. J. Pharm. 2010, 390, 198–207. [Google Scholar] [CrossRef]
- Park, H.S.; Jung, H.Y.; Park, E.Y.; Kim, J.; Lee, W.J.; Bae, Y.S. Cutting edge: Direct interaction of TLR4 with NAD(P)H oxidase 4 isozyme is essential for lipopolysaccharide-induced production of reactive oxygen species and activation of NF-kappa B. J. Immunol. 2004, 173, 3589–3593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soenen, S.J.H.; Illyes, E.; Vercauteren, D.; Braeckmans, K.; Majer, Z.; De Smedt, S.C.; De Cuyper, M. The role of nanoparticle concentration-dependent induction of cellular stress in the internalization of non-toxic cationic magnetoliposomes. Biomaterials 2009, 30, 6803–6813. [Google Scholar] [CrossRef]
- Shi, J.; Yu, S.; Zhu, J.; Zhi, D.; Zhao, Y.; Cui, S.; Zhang, S. Carbamate-linked cationic lipids with different hydrocarbon chains for gene delivery. Colloids Surf. B Biointerfaces 2016, 141, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Habib, S.; Daniels, A.; Ariatti, M.; Singh, M. Anti-c-myc cholesterol based lipoplexes as onco-nanotherapeutic agents in vitro. F1000Research 2020, 9, 770. [Google Scholar] [CrossRef] [PubMed]
- Bulbake, U.; Kommineni, N.; Ionov, M.; Bryszewska, M.; Khan, W. Comparison of cationic liposome and PAMAM dendrimer for delivery of anti-Plk1siRNA in breast cancer treatment. Pharm. Dev. Technol. 2020, 25, 9–19. [Google Scholar]
- Zhang, L.; Mu, C.; Zhang, T.; Wang, Y.; Wang, Y.; Fan, L.; Liu, C.; Chen, H.; Shen, J.; Wei, K.; et al. Systemic Delivery of Aptamer-Conjugated XBP1 siRNA Nanoparticles for Efficient Suppression of HER2+ Breast Cancer. ACS Appl. Mater Interfaces 2020, 12, 32360–32371. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Park, Y.H. Trastuzumab deruxtecan for HER2+ advanced breast cancer. Future Oncol. 2022, 18, 7–19. [Google Scholar] [CrossRef]







| Liposomal Formulation | Molar Ratios of the Respective Cationic Liposome Components (μmol/500 μL) | Total Lipid Content (µg/µL) | ||
|---|---|---|---|---|
| Cytofectin | DOPE | PEG | ||
| Chol-T: DOPE | 1.00 | 1.00 | - | 2.517 |
| Chol-T:DOPE:2% PEG | 1.00 | 0.96 | 0.04 | 2.648 |
| Chol-T:DOPE:5% PEG | 1.00 | 0.90 | 0.10 | 2.938 |
| MS09:DOPE | 1.00 | 1.00 | - | 2.746 |
| MS09:DOPE:2% PEG | 1.00 | 0.96 | 0.04 | 2.914 |
| MS09:DOPE:5% PEG | 1.00 | 0.90 | 0.10 | 3.168 |
| Components | Mass (µg) | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Chol-T:DOPE | 0 | 3.20 | 3.52 | 3.84 | 4.16 | 4.48 * | 4.80 | 5.12 |
| Chol-T:DOPE:2% PEG | 0 | 6.08 | 6.40 | 6.72 | 7.04 | 7.36 | 7.68 * | 8.00 |
| Chol-T:DOPE:5% PEG | 0 | 10.56 | 10.88 | 11.20 | 11.52 | 11.84 * | 12.16 | 12.48 |
| MS09:DOPE | 0 | 5.44 | 5.76 | 6.08 | 6.40 | 6.72 * | 7.04 | 7.36 |
| MS09:DOPE:2% PEG | 0 | 6.08 | 6.40 | 6.72 | 7.04 | 7.36 | 7.68 | 8.00 * |
| MS09:DOPE:5% PEG | 0 | 9.60 | 9.92 | 10.24 | 10.56 | 10.88 | 11.20 | 11.52 * |
| Liposomal Formulation | Sub-Optimum Ratio (w/w) | Optimum Ratio (w/w) | Supra-Optimum Ratio (w/w) |
|---|---|---|---|
| Chol-T | 1:12 | 1:14 | 1:16 |
| Chol-T 2% PEG2000 | 1:22 | 1:24 | 1:26 |
| Chol-T 5% PEG2000 | 1:35 | 1:37 | 1:39 |
| MS09 | 1:19 | 1:21 | 1:23 |
| MS09 2% PEG2000 | 1:23 | 1:25 | 1:27 |
| MS09 5% PEG2000 | 1:34 | 1:36 | 1:38 |
| Cationic Liposome | Liposome | siRNA Lipoplex | ||||
|---|---|---|---|---|---|---|
| Size (nm) | ζ Potential (mV) ± SD | PDI | Size (nm) | ζ Potential (mV) ± SD | PDI | |
| Chol-T | 121.07 ± 11.8 | 44.09 ± 10.56 | 0.321 | 187.97 ± 12.9 | 47.26 ± 5.39 | 0.127 |
| Chol-T 2% PEG | 71.64 ± 2.6 | 32.51 ± 11.02 | 0.304 | 149.61 ± 10.5 | 39.05 ± 8.16 | 0.227 |
| Chol-T 5% PEG | 74.18 ± 1.9 | –1.12 ± 4.818 | 0.136 | 153.24 ± 18.9 | 9.78 ± 1.13 | 0.109 |
| MS09 | 113.02 ± 13.5 | 53.21 ± 4.329 | 0.348 | 169.13 ± 19.2 | 44.61 ± 7.56 | 0.326 |
| MS09 2% PEG | 66.68 ± 1.7 | 39.43 ± 1.185 | 0.136 | 103.24 ± 9.1 | 20.88 ± 3.052 | 0.218 |
| MS09 5% PEG | 65.47 ±1.3 | 16.08 ± 3.799 | 0.269 | 138.64 ± 10.2 | 41.26 ± 9.79 | 0.337 |
| Cationic Liposome | siRNA: Cationic Liposome Endpoint (w/w) | NP Charge Ratio (+/−) |
|---|---|---|
| Chol-T | 1:14 | 1:3.9 |
| Chol-T 2% PEG2000 | 1:24 | 1:6.3 |
| Chol-T 5% PEG2000 | 1:37 | 1:8.8 |
| MS09 | 1:21 | 1:5.4 |
| MS09 2% PEG2000 | 1:25 | 1:6.0 |
| MS09 5% PEG2000 | 1:36 | 1:7.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balgobind, A.; Daniels, A.; Ariatti, M.; Singh, M. HER2/neu Oncogene Silencing in a Breast Cancer Cell Model Using Cationic Lipid-Based Delivery Systems. Pharmaceutics 2023, 15, 1190. https://doi.org/10.3390/pharmaceutics15041190
Balgobind A, Daniels A, Ariatti M, Singh M. HER2/neu Oncogene Silencing in a Breast Cancer Cell Model Using Cationic Lipid-Based Delivery Systems. Pharmaceutics. 2023; 15(4):1190. https://doi.org/10.3390/pharmaceutics15041190
Chicago/Turabian StyleBalgobind, Adhika, Aliscia Daniels, Mario Ariatti, and Moganavelli Singh. 2023. "HER2/neu Oncogene Silencing in a Breast Cancer Cell Model Using Cationic Lipid-Based Delivery Systems" Pharmaceutics 15, no. 4: 1190. https://doi.org/10.3390/pharmaceutics15041190