Adjusting Heterodimeric Coiled-Coils (K/E Zipper) to Connect Autophagy-Inducing Peptide with Cell-Penetrating Peptide
Abstract
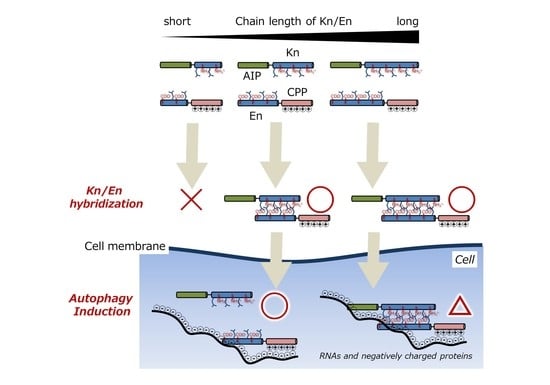
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Antibodies
2.3. Peptide Synthesis
2.4. Fluorescence Titration Curves
2.5. Cell Culture
2.6. Preparation of the K/E Zippers
2.7. Evaluation of the K/E Hybrid Delivery into Cells
2.8. Analysis of Cellular Autophagic Activity
2.9. Cytotoxicity Assay
3. Results and Discussion
3.1. Chemical Structures of Fluorescently Labeled Hetrodimeric Coiled-Coil Modified with AIP or CPP
3.2. Comparison of Binding Constant of the AIP-Kn/En-CPP Hybrids
3.3. Intracellular Delivery of AIP-Kn/En-CPP Hybrids
3.4. Evaluation of Autophagy Induction by the Hybrid of AIP-Kn and En-CPP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Futaki, S.; Hirose, H.; Nakase, I. Arginine-rich peptides: Methods of translocation through biological membranes. Curr. Pharm. Des. 2013, 19, 2863–2868. [Google Scholar] [CrossRef] [PubMed]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-penetrating peptides: From basic research to clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, Y.; Zhang, X.; Zhang, W.; Guo, S.; Jin, F. Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J. Control. Release 2014, 174, 126–136. [Google Scholar] [CrossRef]
- Bolhassani, A.; Jafarzade, B.S.; Mardani, G. In vitro and in vivo delivery of therapeutic proteins using cell penetrating peptides. Peptides 2017, 87, 50–63. [Google Scholar] [CrossRef]
- Borrelli, A.; Tornesello, A.L.; Tornesello, M.L.; Buonaguro, F.M. Cell penetrating peptides as molecular carriers for anti-cancer agents. Molecules 2018, 23, 295–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Copolovici, D.M.; Langel, K.; Eriste, E.; Langel, Ü. Cell-penetrating peptides: Design, synthesis, and applications. ACS Nano 2014, 8, 1972–1994. [Google Scholar] [CrossRef] [PubMed]
- Hitsuda, T.; Michiue, H.; Kitamatsu, M.; Fujimura, A.; Wang, F.; Yamamoto, T.; Han, X.-J.; Tazawa, H.; Uneda, A.; Ohmori, I.; et al. A protein transduction method using oligo-arginine (3R) for the delivery of transcription factors into cell nuclei. Biomaterials 2012, 33, 4665–4672. [Google Scholar] [CrossRef] [PubMed]
- Altrichter, Y.; Seitz, O. Simultaneous targeting of two master regulators of apoptosis with dual-action PNA− and DNA−peptide conjugates. Bioconjugate Chem. 2020, 31, 1928–1937. [Google Scholar] [CrossRef]
- Yu, S.; Yang, H.; Li, T.; Pan, H.; Ren, S.; Luo, G.; Jiang, J.; Yu, L.; Chen, B.; Zhang, Y.; et al. Efficient intracellular delivery of proteins by a multifunctional chimaeric peptide in vitro and in vivo. Nat. Commun. 2021, 12, 5131–5143. [Google Scholar] [CrossRef]
- June, R.K.; Gogoi, K.; Eguchi, K.A.; Cui, X.-S.; Dowdy, S.F. Synthesis of a pH-sensitive nitrilotriacetic linker to peptide transduction domains to enable intracellular delivery of histidine imidazole ring-containing macromolecules. J. Am. Chem. Soc. 2010, 132, 10680–10682. [Google Scholar] [CrossRef]
- Hansen, M.B.; Verdurmen, W.P.R.; Leunissen, E.H.P.; Minten, I.; van Hest, J.C.M.; Brock, R.; Löwik, D.W.P.M. A modular and noncovalent transduction system for leucine-zipper-tagged proteins. ChemBioChem 2011, 12, 2294–2297. [Google Scholar] [CrossRef] [PubMed]
- Rhys, G.G.; Cross, J.A.; Dawson, W.M.; Thompson, H.F.; Shanmugaratnam, S.; Savery, N.J.; Dodding, M.P.; Höcker, B.; Woolfson, D.N. De novo designed peptides for cellular delivery and subcellular localization. Nat. Chem. Biol. 2022, 18, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.F.L.; Wallabregue, A.L.D.; Franz, L.; Hackenberger, C.P.R. Targeted subcellular protein delivery using cleavable cyclic cell-penetrating peptides. Bioconjugate Chem. 2019, 30, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Gui, W.; Ott, C.A.; Yang, K.; Chung, J.S.; Shen, S.; Zhuang, Z. Cell-permeable activity-based ubiquitin probes enable intracellular profiling of human deubiquitinases. J. Am. Chem. Soc. 2018, 140, 12424–12433. [Google Scholar] [CrossRef] [PubMed]
- Herce, H.D.; Schumacher, D.; Schneider, A.F.L.; Ludwig, A.K.; Mann, F.A.; Fillies, M.; Kasper, M.-A.; Reinke, S.; Krause, E.; Leonhardt, H.; et al. Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat. Chem. 2017, 9, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Hakata, Y.; Tsuchiya, S.; Michiue, H.; Ohtsuki, T.; Matsui, H.; Miyazawa, M.; Kitamatsu, M. A novel leucine zipper motif-based hybrid peptide delivers a functional peptide cargo inside cells. Chem. Commun. 2015, 51, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Hakata, Y.; Michiue, H.; Ohtsuki, T.; Miyazawa, M.; Kitamatsu, M. A leucine zipper-based peptide hybrid delivers functional nanog protein inside the cell nucleus. Bioorg. Med. Chem. Lett. 2019, 29, 878–881. [Google Scholar] [CrossRef]
- Hakata, Y.; Ishikawa, S.; Ohtsuki, T.; Miyazawa, M.; Kitamatsu, M. Intracellular delivery of a peptide nucleic acid-based hybrid of an autophagy inducing peptide with a cell-penetrating peptide. Org. Biomol. Chem. 2020, 18, 1978–1986. [Google Scholar] [CrossRef]
- Kitamatsu, M.; Yuasa, H.; Ohtsuki, T.; Michiue, H. Complementary leucine zippering system for effective intracellular delivery of proteins by cell-penetrating peptides. Bioorg. Med. Chem. 2021, 33, 116036–116042. [Google Scholar] [CrossRef]
- Wendt, H.; Leder, L.; Härmä, H.; Jelesarov, I.; Baici, A.; Bosshard, H.R. Very rapid, ionic strength-dependent association and folding of a heterodimeric leucine zipper. Biochemistry 1997, 36, 204–213. [Google Scholar] [CrossRef]
- Dürr, E.; Jelesarov, I.; Bosshard, H.R. Extremely fast folding of a very stable leucine zipper with a strengthened hydrophobic core and lacking electrostatic interactions between helices. Biochemistry 1999, 38, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Shoji-Kawata, S.; Sumpter, R., Jr.; Leveno, M.; Campbell, G.R.; Zou, Z.; Kinch, L.; Wilkins, A.D.; Sun, Q.; Pallauf, K.; MacDuff, D.; et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 2013, 494, 201–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Michiue, H.; Tomizawa, K.; Wei, F.-Y.; Matsushita, M.; Lu, Y.-F.; Ichikawa, T.; Tamiya, T.; Date, I.; Matsui, H. The NH2 terminus of influenza virus hemagglutinin-2 subunit peptides enhances the antitumor potency of polyarginine-mediated p53 protein transduction. J. Biol. Chem. 2005, 280, 8285–8289. [Google Scholar] [CrossRef] [Green Version]
- Barras, D.; Widmann, C. Promises of apoptosis-inducing peptides in cancer therapeutics. Curr. Pharm. Biotechnol. 2011, 12, 1153–1165. [Google Scholar] [CrossRef]
- Litowski, J.R.; Hodges, R.S. Designing heterodimeric two-stranded α-helical coiled-coils. J. Biol. Chem. 2002, 277, 37272–37279. [Google Scholar] [CrossRef] [Green Version]
- Bode, S.A.; Kruis, I.C.; Adams, H.P.J.H.M.; Boelens, W.C.; Pruijn, G.J.M.; Van Hest, J.C.M.; Löwik, D.W.P.M. Coiled-coil-mediated activation of oligoarginine cell-penetrating peptides. ChemBioChem 2017, 18, 185–188. [Google Scholar] [CrossRef] [Green Version]
- Mikolajczak, D.J.; Heier, J.L.; Schade, B.; Koksch, B. Catalytic activity of peptide−nanoparticle conjugates regulated by a conformational change. Biomacromolecules 2017, 18, 3557–3562. [Google Scholar] [CrossRef]
- Yano, Y.; Yano, A.; Oishi, S.; Sugimoto, Y.; Tsujimoto, G.; Fujii, N.; Matsuzaki, K. Coiled-coil tag-probe system for quick labeling of membrane receptors in living cells. ACS Chem. Biol. 2008, 3, 341–345. [Google Scholar] [CrossRef]
- Daudey, G.A.; Shen, M.; Singhal, A.; van der Est, P.; Sevink, G.J.A.; Boyle, A.L.; Kros, A. Liposome fusion with orthogonal coiled coil peptides as fusogens: The efficacy of roleplaying peptides. Chem. Sci. 2021, 12, 13782–13792. [Google Scholar] [CrossRef]
- Crone, N.S.A.; Kros, A.; Boyle, A.L. Modulation of coiled-coil binding strength and fusogenicity through peptide stapling. Bioconjugate Chem. 2020, 31, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, A.; Watanabe, S.; Goel, I.; Ishihara, K.; Ekdahl, K.N.; Nilsson, B.; Teramura, Y. Promotion of cell membrane fusion by cell-cell attachment through cell surface modification with functional peptide-PEG-lipids. Biomaterials 2020, 253, 120113–120122. [Google Scholar] [CrossRef] [PubMed]
- Peraro, L.; Zou, Z.; Makwana, K.M.; Cummings, A.E.; Ball, H.L.; Yu, H.; Lin, Y.; Levine, B.; Kritzer, J.A. Diversity-oriented stapling yields intrinsically cell-penetrant inducers of autophagy. J. Am. Chem. Soc. 2017, 139, 7792–7802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heitz, F.; Morris, M.C.; Fesquet, D.; Cavadore, J.C.; Dorée, M.; Divita, G. Interactions of cyclins with cyclin-dependent kinases: A common interactive mechanism. Biochemistry 1997, 36, 4995–5003. [Google Scholar] [CrossRef] [PubMed]
- Sgolastra, F.; Backlund, C.M.; Ozay, E.I.; deRonde, B.M.; Minter, L.M.; Tew, G.N. Sequence segregation improves non-covalent protein delivery. J. Control. Release 2017, 254, 131–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yagi, H.; Tomono, T.; Handa, Y.; Saito, N.; Ukawa, M.; Miyata, K.; Shigeno, K.; Sakuma, S. Performance of cell-penetrating peptides anchored to polysaccharide platforms applied via various mucosal routes as an absorption enhancer. Mol. Pharm. 2023, 20, 303–313. [Google Scholar] [CrossRef]
- Wu, Y.; Jiang, W.; Cong, Z.; Chen, K.; She, Y.; Zhong, C.; Zhang, W.; Chen, M.; Zhou, M.; Shao, N.; et al. An effective strategy to develop potent and selective antifungal agents from cell penetrating peptides in tackling drug-resistant invasive fungal infections. J. Med. Chem. 2022, 65, 7296–7311. [Google Scholar] [CrossRef]
- Bolaños, K.; Sánchez-Navarro, M.; Tapia-Arellan, A.; Giralt, E.; Kogan, M.J.; Araya, E. Oligoarginine peptide conjugated to BSA improves cell penetration of gold nanorods and nanoprisms for biomedical applications. Pharmaceutics 2021, 13, 1204–1219. [Google Scholar] [CrossRef]
- Vega-Rubín-de-Celis, S.; Zou, Z.; Fernández, Á.F.; Ci, B.; Kim, M.; Xiao, G.; Xie, Y.; Levine, B. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc. Natl. Acad. Sci. USA 2018, 115, 4176–4181. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zou, Z.; Sun, D.; Li, Y.; Sinha, S.C.; Yu, L.; Bennett, L.; Levine, B. GLIPR2 is a negative regulator of autophagy and the BECN1-ATG14-containing phosphatidylinositol 3-kinase complex. Autophagy 2021, 17, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Gao, Y.; Cheng, Y. A manganese (II)-based coordinative dendrimer with robust efficiency in intracellular peptide delivery. Bioact. Mater. 2022, 9, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.T.; Boyle, A.; Marsden, H.R.; Valdink, D.; Martelli, G.; Raap, J.; Kros, A. Probing coiled-coil assembly by paramagnetic NMR spectroscopy. Org. Biomol. Chem. 2015, 13, 1159–1168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, P.; van Son, M.; Zheng, T.; Valdink, D.; Raap, J.; Kros, A.; Huber, M. Coiled-coil formation of the membrane-fusion K/E peptides viewed by electron paramagnetic resonance. PLoS ONE 2018, 13, e0191197. [Google Scholar] [CrossRef] [Green Version]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hakata, Y.; Yamashita, K.; Hashimoto, S.; Ohtsuki, T.; Miyazawa, M.; Kitamatsu, M. Adjusting Heterodimeric Coiled-Coils (K/E Zipper) to Connect Autophagy-Inducing Peptide with Cell-Penetrating Peptide. Pharmaceutics 2023, 15, 1048. https://doi.org/10.3390/pharmaceutics15041048
Hakata Y, Yamashita K, Hashimoto S, Ohtsuki T, Miyazawa M, Kitamatsu M. Adjusting Heterodimeric Coiled-Coils (K/E Zipper) to Connect Autophagy-Inducing Peptide with Cell-Penetrating Peptide. Pharmaceutics. 2023; 15(4):1048. https://doi.org/10.3390/pharmaceutics15041048
Chicago/Turabian StyleHakata, Yoshiyuki, Kazuma Yamashita, Sonoko Hashimoto, Takashi Ohtsuki, Masaaki Miyazawa, and Mizuki Kitamatsu. 2023. "Adjusting Heterodimeric Coiled-Coils (K/E Zipper) to Connect Autophagy-Inducing Peptide with Cell-Penetrating Peptide" Pharmaceutics 15, no. 4: 1048. https://doi.org/10.3390/pharmaceutics15041048