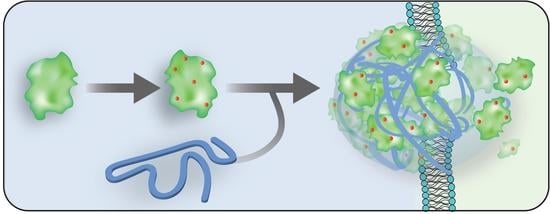
Direct Cytosolic Delivery of Citraconylated Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
- Nanocomposite preparation, characterization, and instrumentation
- Confocal microscopy
- Imaging flow cytometry
- Transmission Electron Microscopy (TEM)
- Protein expression
3. Results and Discussion
3.1. Modification of EGFP with CA and Verification of Impact of Modification on Proteins
Verification Impact of the Modification on Proteins
3.2. Fabrication and Characterization of PONI Guan/c−EGFP Conjugates
3.3. Cytosolic Protein Delivery, and Nuclear Access of EGFP
3.4. Delivery of c−EGFP to Different Cell Lines and Cytosolic Delivery of Different Citraconylated Proteins
3.5. Delivery of Functional Enzyme Cre Recombinase A and RNase A
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scaletti, F.; Hardie, J.; Lee, Y.-W.; Luther, D.C.; Ray, M.; Rotello, V.M. Protein Delivery into Cells Using Inorganic Nanoparticle–Protein Supramolecular Assemblies. Chem. Soc. Rev. 2018, 47, 3421–3432. [Google Scholar] [CrossRef] [PubMed]
- Vargason, A.M.; Anselmo, A.C.; Mitragotri, S. The Evolution of Commercial Drug Delivery Technologies. Nat. Biomed. Eng. 2021, 5, 951–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Song, W.; Lu, Y.; Xu, Y.; Wang, C.; Yu, D.G.; Kim, I. Recent Advances in Poly(α-L-Glutamic Acid)-Based Nanomaterials for Drug Delivery. Biomolecules 2022, 12, 636. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Varyambath, A.; Ding, Y.; Chen, B.; Huang, X.; Zhang, Y.; Yu, D.; Kim, I.; Song, W. Porous Organic Polymers for Drug Delivery: Hierarchical Pore Structures, Variable Morphologies, and Biological Properties. Biomater. Sci. 2022, 10, 5369–5390. [Google Scholar] [CrossRef]
- Luther, D.C.; Jeon, T.; Goswami, R.; Nagaraj, H.; Kim, D.; Lee, Y.-W.; Rotello, V.M. Protein Delivery: If Your GFP (or Other Small Protein) Is in the Cytosol, It Will Also Be in the Nucleus. Bioconjug. Chem. 2021, 32, 891–896. [Google Scholar] [CrossRef]
- Raman, V.; Van Dessel, N.; Hall, C.L.; Wetherby, V.E.; Whitney, S.A.; Kolewe, E.L.; Bloom, S.M.K.; Sharma, A.; Hardy, J.A.; Bollen, M.; et al. Intracellular Delivery of Protein Drugs with an Autonomously Lysing Bacterial System Reduces Tumor Growth and Metastases. Nat. Commun. 2021, 12, 6116. [Google Scholar] [CrossRef]
- Lee, Y.-W.; Luther, D.C.; Kretzmann, J.A.; Burden, A.; Jeon, T.; Zhai, S.; Rotello, V.M. Protein Delivery into the Cell Cytosol Using Non-Viral Nanocarriers. Theranostics 2019, 9, 3280–3292. [Google Scholar] [CrossRef]
- Goswami, R.; Jeon, T.; Nagaraj, H.; Zhai, S.; Rotello, V.M. Accessing Intracellular Targets through Nanocarrier-Mediated Cytosolic Protein Delivery. Trends Pharmacol. Sci. 2020, 41, 743–754. [Google Scholar] [CrossRef]
- Luther, D.C.; Huang, R.; Jeon, T.; Zhang, X.; Lee, Y.-W.; Nagaraj, H.; Rotello, V.M. Delivery of Drugs, Proteins, and Nucleic Acids Using Inorganic Nanoparticles. Adv. Drug Deliv. Rev. 2020, 156, 188–213. [Google Scholar] [CrossRef]
- Teo, S.L.Y.; Rennick, J.J.; Yuen, D.; Al-Wassiti, H.; Johnston, A.P.R.; Pouton, C.W. Unravelling Cytosolic Delivery of Cell Penetrating Peptides with a Quantitative Endosomal Escape Assay. Nat. Commun. 2021, 12, 3721. [Google Scholar] [CrossRef]
- Smith, S.A.; Selby, L.I.; Johnston, A.P.R.; Such, G.K. The Endosomal Escape of Nanoparticles: Toward More Efficient Cellular Delivery. Bioconjug. Chem. 2019, 30, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Lau, S.Y.; Lim, Z.W.; Chang, S.C.; Ghadessy, F.; Partridge, A.; Miserez, A. Phase-Separating Peptides for Direct Cytosolic Delivery and Redox-Activated Release of Macromolecular Therapeutics. Nat. Chem. 2022, 14, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Herce, H.D.; Schumacher, D.; Schneider, A.F.L.; Ludwig, A.K.; Mann, F.A.; Fillies, M.; Kasper, M.A.; Reinke, S.; Krause, E.; Leonhardt, H.; et al. Cell-Permeable Nanobodies for Targeted Immunolabelling and Antigen Manipulation in Living Cells. Nat. Chem. 2017, 9, 762–771. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Liew, S.S.; Li, L.; Yao, S.Q. Bypassing Endocytosis: Direct Cytosolic Delivery of Proteins. J. Am. Chem. Soc. 2018, 140, 15986–15996. [Google Scholar] [CrossRef] [PubMed]
- Mout, R.; Ray, M.; Tay, T.; Sasaki, K.; Yesilbag Tonga, G.; Rotello, V.M. General Strategy for Direct Cytosolic Protein Delivery via Protein-Nanoparticle Co-Engineering. ACS Nano 2017, 11, 6416–6421. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.W.; Luther, D.C.; Goswami, R.; Jeon, T.; Clark, V.; Elia, J.; Gopalakrishnan, S.; Rotello, V.M. Direct Cytosolic Delivery of Proteins through Coengineering of Proteins and Polymeric Delivery Vehicles. J. Am. Chem. Soc. 2020, 142, 4349–4355. [Google Scholar] [CrossRef]
- Luther, D.C.; Lee, Y.W.; Nagaraj, H.; Clark, V.; Jeon, T.; Goswami, R.; Gopalakrishnan, S.; Fedeli, S.; Jerome, W.; Elia, J.L.; et al. Cytosolic Protein Delivery Using Modular Biotin-Streptavidin Assembly of Nanocomposites. ACS Nano 2022, 16, 7323–7330. [Google Scholar] [CrossRef]
- Lee, Y.; Ishii, T.; Cabral, H.; Kim, H.J.; Seo, J.H.; Nishiyama, N.; Oshima, H.; Osada, K.; Kataoka, K. Charge-Conversional Polyionic Complex Micelles-Efficient Nanocarriers for Protein Delivery into Cytoplasm. Angew. Chemie-Int. Ed. 2009, 48, 5309–5312. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ishii, T.; Kim, H.J.; Nishiyama, N.; Hayakawa, Y.; Itaka, K.; Kataoka, K. Efficient Delivery of Bioactive Antibodies into the Cytoplasm of Living Cells by Charge-Conversional Polyion Complex Micelles. Angew. Chemie–Int. Ed. 2010, 49, 2552–2555. [Google Scholar] [CrossRef]
- Xie, J.; Gonzalez-Carter, D.; Tockary, T.A.; Nakamura, N.; Xue, Y.; Nakakido, M.; Akiba, H.; Dirisala, A.; Liu, X.; Toh, K.; et al. Dual-Sensitive Nanomicelles Enhancing Systemic Delivery of Therapeutically Active Antibodies Specifically into the Brain. ACS Nano 2020, 14, 6729–6742. [Google Scholar] [CrossRef]
- Chen, P.; Yang, W.; Hong, T.; Miyazaki, T.; Dirisala, A.; Kataoka, K.; Cabral, H. Nanocarriers Escaping from Hyperacidified Endo/Lysosomes in Cancer Cells Allow Tumor-Targeted Intracellular Delivery of Antibodies to Therapeutically Inhibit c-MYC. Biomaterials 2022, 288, 121748. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Kim, Y.; Song, Y.; Choi, J.U.; Park, E.; Choi, W.; Park, J.; Lee, Y. Comparison of PH-Sensitive Degradability of Maleic Acid Amide Derivatives. Bioorganic Med. Chem. Lett. 2014, 24, 2364–2367. [Google Scholar] [CrossRef] [PubMed]
- Arndt, C.; Koristka, S.; Feldmann, A.; Bachmann, M. Native Polyacrylamide Gels. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2019; Volume 1855, pp. 87–91. [Google Scholar]
- Buecheler, J.W.; Winzer, M.; Weber, C.; Gieseler, H. Alteration of Physicochemical Properties for Antibody-Drug Conjugates and Their Impact on Stability. J. Pharm. Sci. 2020, 109, 161–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasso, L.; Purdie, L.; Grabowska, A.; Jones, A.T.; Alexander, C. Time and Cell-Dependent Effects of Endocytosis Inhibitors on the Internalization of Biomolecule Markers and Nanomaterials. J. Interdiscip. Nanomed. 2018, 3, 67–81. [Google Scholar] [CrossRef]
- Lin, H.P.; Singla, B.; Ghoshal, P.; Faulkner, J.L.; Cherian-Shaw, M.; O’Connor, P.M.; She, J.X.; Belin de Chantemele, E.J.; Csányi, G. Identification of Novel Macropinocytosis Inhibitors Using a Rational Screen of Food and Drug Administration-Approved Drugs. Br. J. Pharmacol. 2018, 175, 3640–3655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahammad, S.; Parmryd, I. Cholesterol Depletion Using Methyl-β-Cyclodextrin. Methods Mol. Biol. 2015, 1232, 91–102. [Google Scholar] [PubMed] [Green Version]
- Fu, J.; Yu, C.; Li, L.; Yao, S.Q. Intracellular Delivery of Functional Proteins and Native Drugs by Cell-Penetrating Poly(Disulfide)S. J. Am. Chem. Soc. 2015, 137, 12153–12160. [Google Scholar] [CrossRef]
- Pan, R.; Xu, W.; Ding, Y.; Lu, S.; Chen, P. Uptake Mechanism and Direct Translocation of a New CPP for SiRNA Delivery. Mol. Pharm. 2016, 13, 1366–1374. [Google Scholar] [CrossRef]
- Wang, R.; Brattain, M.G. The Maximal Size of Protein to Diffuse through the Nuclear Pore Is Larger than 60 KDa. FEBS Lett. 2007, 581, 3164–3170. [Google Scholar] [CrossRef] [Green Version]
- Nagy, A. Cre Recombinase: The Universal Reagent for Genome Tailoring. Genesis 2000, 26, 99–109. [Google Scholar] [CrossRef]
- Sternberg, N.; Hamilton, D. Bacteriophage P1 Site-Specific Recombination. I. Recombination between LoxP Sites. J. Mol. Biol. 1981, 150, 467–486. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Duan, F.; Liu, S.; Wu, T.; Shang, Y.; Tian, R.; Liu, J.; Wang, Z.G.; Jiang, Q.; Ding, B. Efficient Intracellular Delivery of RNase A Using DNA Origami Carriers. ACS Appl. Mater. Interfaces 2019, 11, 11112–11118. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Li, Q.; Neufeld, C.I.; Pouli, D.; Sun, S.; Yang, L.; Deng, P.; Wang, M.; Georgakoudi, I.; et al. Hyaluronic Acid Modification of RNase A and Its Intracellular Delivery Using Lipid-like Nanoparticles. J. Control Release 2017, 263, 39–45. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goswami, R.; Lehot, V.; Çiçek, Y.A.; Nagaraj, H.; Jeon, T.; Nguyen, T.; Fedeli, S.; Rotello, V.M. Direct Cytosolic Delivery of Citraconylated Proteins. Pharmaceutics 2023, 15, 218. https://doi.org/10.3390/pharmaceutics15010218
Goswami R, Lehot V, Çiçek YA, Nagaraj H, Jeon T, Nguyen T, Fedeli S, Rotello VM. Direct Cytosolic Delivery of Citraconylated Proteins. Pharmaceutics. 2023; 15(1):218. https://doi.org/10.3390/pharmaceutics15010218
Chicago/Turabian StyleGoswami, Ritabrita, Victor Lehot, Yağız Anıl Çiçek, Harini Nagaraj, Taewon Jeon, Terry Nguyen, Stefano Fedeli, and Vincent M. Rotello. 2023. "Direct Cytosolic Delivery of Citraconylated Proteins" Pharmaceutics 15, no. 1: 218. https://doi.org/10.3390/pharmaceutics15010218