Tuning Design Parameters of ICAM-1-Targeted 3DNA Nanocarriers to Optimize Pulmonary Targeting Depending on Drug Type
Abstract
:1. Introduction
2. Materials and Methods
2.1. Antibodies and Reagents
2.2. Preparation of Anti-ICAM/3DNA NCs
2.3. 125Iodine Labeling of Antibody-Oligonucleotide Conjugates
2.4. Biodistribution of Anti-ICAM/3DNA in Mice
2.5. Visualization of Lung Targeting of Anti-ICAM/3DNA in Mice
2.6. Ethical Use of Laboratory Animals
2.7. Statistical Analysis
3. Results and Discussion
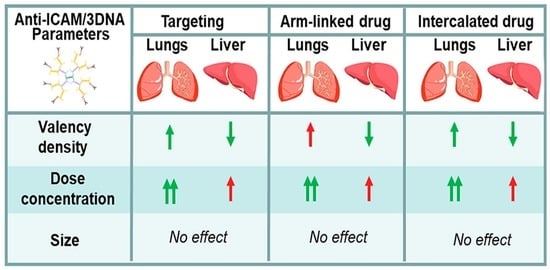
3.1. Role of Targeting Valency and Dose Concentration on the Biodistribution of 4-Layer Anti-ICAM/3DNA
3.2. Significance of Targeting Valency and Dose Concentration of 4L Anti-ICAM/3DNA for Intercalating vs. Arm-Linked Drugs
3.3. Role of Targeting Valency and Dose Concentration on the Biodistribution of 2-Layer Anti-ICAM/3DNA
3.4. Significance of Targeting Valency and Dose Concentration of 2L Anti-ICAM/3DNA for Intercalating vs. Arm-Linked Drugs
3.5. Multiparametric Comparison between 2L and 4L Anti-ICAM/3DNA Biodistribution
3.6. Comparative Drug Delivery Capacity for 2L and 4L anti-ICAM/3DNA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Getts, R.; Muro, S. DNA-Based Drug Carriers: The Paradox of a Classical “Cargo” Material Becoming a Versatile “Carrier” to Overcome Barriers in Drug Delivery. Curr. Pharm. Des. 2016, 22, 1245–1258. [Google Scholar] [CrossRef]
- Lee, H.; Lytton-Jean, A.K.R.; Chen, Y.; Love, K.T.; Park, A.I.; Karagiannis, E.D.; Sehgal, A.; Querbes, W.; Zurenko, C.S.; Jayaraman, M.; et al. Molecularly Self-Assembled Nucleic Acid Nanoparticles for Targeted in Vivo siRNA Delivery. Nat. Nanotechnol. 2012, 7, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Schüller, V.J.; Heidegger, S.; Sandholzer, N.; Nickels, P.C.; Suhartha, N.A.; Endres, S.; Bourquin, C.; Liedl, T. Cellular Immunostimulation by CpG-Sequence-Coated DNA Origami Structures. ACS Nano 2011, 5, 9696–9702. [Google Scholar] [CrossRef] [Green Version]
- Bagalkot, V.; Farokhzad, O.C.; Langer, R.; Jon, S. An Aptamer-Doxorubicin Physical Conjugate as a Novel Targeted Drug-Delivery Platform. Angew. Chem. Int. Ed. 2006, 45, 8149–8152. [Google Scholar] [CrossRef] [PubMed]
- Rosi, N.L.; Giljohann, D.A.; Thaxton, C.S.; Lytton-Jean, A.K.R.; Han, M.S.; Mirkin, C.A. Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science 2006, 312, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Gerhart, J.; Greenbaum, M.; Casta, L.; Clemente, A.; Mathers, K.; Getts, R.; George-Weinstein, M. Antibody-Conjugated, DNA-Based Nanocarriers Intercalated with Doxorubicin Eliminate Myofibroblasts in Explants of Human Lens Tissue. J. Pharmacol. Exp. Ther. 2017, 361, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Roki, N.; Tsinas, Z.; Solomon, M.; Bowers, J.; Getts, R.C.; Muro, S. Unprecedently High Targeting Specificity toward Lung ICAM-1 Using 3DNA Nanocarriers. J. Control. Release 2019, 305, 41–49. [Google Scholar] [CrossRef]
- Muro, S. A DNA Device that Mediates Selective Endosomal Escape and Intracellular Delivery of Drugs and Biologicals. Adv. Funct. Mater. 2014, 24, 2899–2906. [Google Scholar] [CrossRef] [Green Version]
- Douglas, S.M.; Bachelet, I.; Church, G.M. A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 2012, 335, 831–834. [Google Scholar] [CrossRef]
- Zhang, K.; Hao, L.; Hurst, S.J.; Mirkin, C.A. Antibody-Linked Spherical Nucleic Acids for Cellular Targeting. J. Am. Chem. Soc. 2012, 134, 16488–16491. [Google Scholar] [CrossRef] [Green Version]
- Chang, M.; Yang, C.-S.; Huang, D.-M. Aptamer-Conjugated DNA Icosahedral Nanoparticles as a Carrier of Doxorubicin for Cancer Therapy. ACS Nano 2011, 5, 6156–6163. [Google Scholar] [CrossRef]
- Bujold, K.E.; Lacroix, A.; Sleiman, H.F. DNA Nanostructures at the Interface with Biology. Chem 2018, 4, 495–521. [Google Scholar] [CrossRef] [Green Version]
- Nilsen, T.W.; Grayzel, J.; Prensky, W. Dendritic Nucleic Acid Structures. J. Theor. Biol. 1997, 187, 273–284. [Google Scholar] [CrossRef]
- Huang, Y.-H.; Peng, W.; Furuuchi, N.; Gerhart, J.V.; Rhodes, K.; Mukherjee, N.; Jimbo, M.; Gonye, G.E.; Brody, J.R.; Getts, R.C.; et al. Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth. Cancer Res. 2016, 76, 1549–1559. [Google Scholar] [CrossRef] [Green Version]
- Mora, J.R.; Getts, R.C. High-Sensitivity Detection Methods for Low-Abundance RNA Species: Applications for functional genomics research. Expert Rev. Mol. Diagn. 2007, 7, 775–785. [Google Scholar] [CrossRef]
- Muro-Galindo, S.; Muzykantov, V.R. Targeted Carriers for Intracellular Drug Delivery. U.S. Patent No. 9,707,299, 18 July 2017. [Google Scholar]
- Sawicki, J.A.; Peng, W.; Rhodes, K.; Getts, R. Abstract 700: Systemic Administration of DNA Nanoparticles Containing the Diphtheria Toxin Gene Reduces Pancreatic Tumor Load in Mice. Cancer Res. 2014, 74, 700. [Google Scholar] [CrossRef]
- Van Steenwinckel, J.; Schang, A.-L.; Krishnan, M.L.; Degos, V.; Delahaye-Duriez, A.; Bokobza, C.; Csaba, Z.; Verdonk, F.; Montané, A.; Sigaut, S.; et al. Decreased Microglial Wnt/β-Catenin Signalling Drives Microglial pro-Inflammatory Activation in the Developing Brain. Brain 2019, 142, 3806–3833. [Google Scholar] [CrossRef]
- Gerhart, J.; Werner, L.; Mamalis, N.; Infanti, J.; Withers, C.; Abdalla, F.; Gerhart, C.; Bravo-Nuevo, A.; Gerhart, O.; Getts, L.; et al. Depletion of Myo/Nog Cells in the Lens Mitigates Posterior Capsule Opacification in Rabbits. Investig. Opthalmol. Vis. Sci. 2019, 60, 1813–1823. [Google Scholar] [CrossRef] [Green Version]
- Muro, S. Intercellular Adhesion Molecule-1 and Vascular Cell Adhesion Molecule-1. In Endothelial Biomedicine; Aird, W.C., Ed.; Cambridge University Press: New York, NY, USA, 2007; pp. 1058–1070. [Google Scholar]
- Garnacho, C.; Dhami, R.; Solomon, M.; Schuchman, E.H.; Muro, S. Enhanced Delivery and Effects of Acid Sphingomyelinase by ICAM-1-Targeted Nanocarriers in Type B Niemann-Pick Disease Mice. Mol. Ther. 2017, 25, 1686–1696. [Google Scholar] [CrossRef] [Green Version]
- Muro, S.; Garnacho, C.; Champion, J.A.; Leferovich, J.; Gajewski, C.; Schuchman, E.H.; Mitragotri, S.; Muzykantov, V.R. Control of Endothelial Targeting and Intracellular Delivery of Therapeutic Enzymes by Modulating the Size and Shape of ICAM-1-targeted Carriers. Mol. Ther. 2008, 16, 1450–1458. [Google Scholar] [CrossRef]
- Weller, G.E.; Villanueva, F.S.; Tom, E.M.; Wagner, W.R. Targeted Ultrasound Contrast Agents: In Vitro Assessment of Endothelial Dysfunction and Multi-Targeting to ICAM-1 and Sialyl Lewisx. Biotechnol. Bioeng. 2005, 92, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-S.; Kim, S.-H.; Cai, Q.-Y.; Kim, S.-Y.; Kim, H.-O.; Lee, H.-J.; Kim, E.-A.; Yoon, S.-E.; Yun, K.-J.; Yoon, K.-H. Inflammation-Specific T 1 Imaging Using Anti-Intercellular Adhesion Molecule 1 Antibody-Conjugated Gadolinium Diethylenetriaminepentaacetic Acid. Mol. Imaging 2007, 6, 75–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolhar, P.; Anselmo, A.C.; Gupta, V.; Pant, K.; Prabhakarpandian, B.; Ruoslahti, E.; Mitragotri, S. Using Shape Effects to Target Antibody-Coated Nanoparticles to Lung and Brain Endothelium. Proc. Natl. Acad. Sci. USA 2013, 110, 10753–10758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcos-Contreras, O.A.; Brenner, J.S.; Kiseleva, R.Y.; Zuluaga-Ramirez, V.; Greineder, C.F.; Villa, C.H.; Hood, E.D.; Myerson, J.W.; Muro, S.; Persidsky, Y.; et al. Combining Vascular Targeting and the Local First Pass Provides 100-Fold Higher Uptake of ICAM-1-Targeted vs. Untargeted Nanocarriers in the Inflamed Brain. J. Control. Release 2019, 301, 54–61. [Google Scholar] [CrossRef]
- Karimi Goftar, M.; Moradi Kor, N.; Moradi Kor, Z. DNA Intercalators and Using Them as Anticancer Drugs. Int. J. Adv. Biol. Biomed. Res. 2014, 2, 811–822. [Google Scholar]
- Atochina, E.N.; Balyasnikova, I.V.; Danilov, S.M.; Granger, D.N.; Fisher, A.B.; Muzykantov, V.R. Immunotargeting of Catalase to ACE or ICAM-1 Protects Perfused Rat Lungs against Oxidative Stress. Am. J. Physiol. Content 1998, 275, L806–L817. [Google Scholar] [CrossRef]
- Murciano, J.-C.; Muro, S.; Koniaris, L.; Christofidou-Solomidou, M.; Harshaw, D.W.; Albelda, S.M.; Granger, D.N.; Cines, D.B.; Muzykantov, V.R. ICAM-Directed Vascular Immunotargeting of Antithrombotic Agents to the Endothelial Luminal Surface. Blood 2003, 101, 3977–3984. [Google Scholar] [CrossRef] [Green Version]
- Dhand, C.; Prabhakaran, M.P.; Beuerman, R.W.; Lakshminarayanan, R.; Dwivedi, N.; Ramakrishna, S. Role of Size of Drug Delivery Carriers for Pulmonary and Intravenous Administration with Emphasis on Cancer Therapeutics and Lung-Targeted Drug Delivery. RSC Adv. 2014, 4, 32673–32689. [Google Scholar] [CrossRef]
- Charoenphol, P.; Bermudez, H. Aptamer-Targeted DNA Nanostructures for Therapeutic Delivery. Mol. Pharm. 2014, 11, 1721–1725. [Google Scholar] [CrossRef]
- Calderon, A.J.; Bhowmick, T.; Leferovich, J.; Burman, B.; Pichette, B.; Muzykantov, V.; Eckmann, D.M.; Muro, S. Optimizing Endothelial Targeting by Modulating the Antibody Density and Particle Concentration of Anti-ICAM Coated Carriers. J. Control. Release 2011, 150, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Dormidontova, E.E. Nanoparticle Targeting Using Multivalent Ligands: Computer Modeling. Soft Matter 2011, 7, 4435–4445. [Google Scholar] [CrossRef]
- Gratton, S.E.; Ropp, P.A.; Pohlhaus, P.D.; Luft, J.C.; Madden, V.J.; Napier, M.E.; DeSimone, J.M. The Effect of Particle Design on Cellular Internalization Pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 11613–11618. [Google Scholar] [CrossRef] [Green Version]
- Bastings, M.M.C.; Anastassacos, F.M.; Ponnuswamy, N.; Leifer, F.G.; Cuneo, G.; Lin, C.; Ingber, D.E.; Ryu, J.H.; Shih, W.M. Modulation of the Cellular Uptake of DNA Origami through Control over Mass and Shape. Nano Lett. 2018, 18, 3557–3564. [Google Scholar] [CrossRef]
- Ko, S.; Liu, H.; Chen, Y.; Mao, C. DNA Nanotubes as Combinatorial Vehicles for Cellular Delivery. Biomacromolecules 2008, 9, 3039–3043. [Google Scholar] [CrossRef] [Green Version]
- Roki, N.; Solomon, M.; Casta, L.; Bowers, J.; Getts, R.C.; Muro, S. A Method to Improve Quantitative Radiotracing-Based Analysis of the in Vivo Biodistribution of Drug Carriers. Bioeng. Transl. Med. 2020, 6, e10208. [Google Scholar] [CrossRef]
- Garnacho, C.; Dhami, R.; Simone, E.; Dziubla, T.; Leferovich, J.; Schuchman, E.H.; Muzykantov, V.; Muro, S. Delivery of Acid Sphingomyelinase in Normal and Niemann-Pick Disease Mice Using Intercellular Adhesion Molecule-1-Targeted Polymer Nanocarriers. J. Pharmacol. Exp. Ther. 2008, 325, 400–408. [Google Scholar] [CrossRef] [Green Version]
- Selzner, N.; Selzner, M.; Odermatt, B.; Tian, Y.; van Rooijen, N.; Clavien, P. ICAM-1 Triggers Liver Regeneration through Leukocyte Recruitment and Kupffer Cell-Dependent Release of TNF-α/IL-6 in Mice. Gastroenterology 2003, 124, 692–700. [Google Scholar] [CrossRef]
- Yusuf-Makagiansar, H.; Anderson, M.E.; Yakovleva, T.V.; Murray, J.; Siahaan, T.J. Inhibition of LFA-1/ICAM-1 and VLA-4/VCAM-1 as a Therapeutic Approach to Inflammation and Autoimmune Diseases. Med. Res. Rev. 2002, 22, 146–167. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Y.; Wu, Y.; Wu, J.; Wang, W.; Wu, Q.; Yuan, Z. The Effects of Ligand Valency and Density on the Targeting Ability of Multivalent Nanoparticles Based on Negatively Charged Chitosan Nanoparticles. Colloids Surf. B Biointerfaces 2018, 161, 508–518. [Google Scholar] [CrossRef]
- Serrano, D.; Manthe, R.L.; Paul, E.; Chadha, R.; Muro, S. How Carrier Size and Valency Modulate Receptor-Mediated Signaling: Understanding the Link between Binding and Endocytosis of ICAM-1-Targeted Carriers. Biomacromolecules 2016, 17, 3127–3137. [Google Scholar] [CrossRef] [Green Version]
- Muro, S.; Dziubla, T.; Qiu, W.; Leferovich, J.; Cui, X.; Berk, E.; Muzykantov, V.R. Endothelial Targeting of High-Affinity Multivalent Polymer Nanocarriers Directed to Intercellular Adhesion Molecule 1. J. Pharmacol. Exp. Ther. 2006, 317, 1161–1169. [Google Scholar] [CrossRef] [Green Version]
- Ansar, M.; Serrano, D.; Papademetriou, I.; Bhowmick, T.K.; Muro, S. Biological Functionalization of Drug Delivery Carriers To Bypass Size Restrictions of Receptor-Mediated Endocytosis Independently from Receptor Targeting. ACS Nano 2013, 7, 10597–10611. [Google Scholar] [CrossRef] [Green Version]
- Carver, L.A.; Schnitzer, J.E. Caveolae: Mining Little Caves for New Cancer Targets. Nat. Cancer 2003, 3, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Solomon, M.; Muro, S. Lysosomal Enzyme Replacement Therapies: Historical Development, Clinical Outcomes, and Future Perspectives. Adv. Drug Deliv. Rev. 2017, 118, 109–134. [Google Scholar] [CrossRef]
- Purdie, L.; Alexander, C.; Spain, S.G.; Magnusson, J.P. Influence of Polymer Size on Uptake and Cytotoxicity of Doxorubicin-Loaded DNA–PEG Conjugates. Bioconjugate Chem. 2016, 27, 1244–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, D.; Berrouschot, J.; Brandt, T.; Hacke, W.; Ferbert, A.; Norris, S.; Polmar, S.; Schäfer, E. Safety, Pharmacokinetics and Biological Activity of Enlimomab (Anti-ICAM-1 Antibody): An Open-Label, Dose Escalation Study in Patients Hospitalized for Acute Stroke. Eur. Neurol. 1998, 40, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Ohto, U.; Shibata, T.; Tanji, H.; Ishida, H.; Krayukhina, E.; Uchiyama, S.; Miyake, K.; Shimizu, T. Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9. Nature 2015, 520, 702–705. [Google Scholar] [CrossRef]
- Appella, D.H. Non-Natural Nucleic Acids for Synthetic Biology. Curr. Opin. Chem. Biol. 2009, 13, 687–696. [Google Scholar] [CrossRef] [Green Version]
- Kosuri, S.; Church, G.M. Large-scale de novo DNA synthesis: Technologies and applications. Nat. Methods 2014, 11, 499–507. [Google Scholar] [CrossRef]
- Nagy, A.; Alhatlani, B. An overview of current COVID-19 vaccine platforms. Comput. Struct. Biotechnol. J. 2021, 19, 2508–2517. [Google Scholar] [CrossRef]
- Zielińska, A.; Carreiró, F.; Oliveira, A.; Neves, A.; Pires, B.; Venkatesh, D.; Durazzo, A.; Lucarini, M.; Eder, P.; Silva, A.; et al. Polymeric nanoparticles: Production, characterization, toxicology and ecotoxicology. Molecules 2020, 25, 3731. [Google Scholar] [CrossRef]










| Formulation | Targeting Ab Valency (Ab/NC) | Targeting Ab Density (Ab/µm2) | Mean Diameter (nm) | PDI | ζ-Potential (mV) |
|---|---|---|---|---|---|
| 4L 3DNA | |||||
| IgG control (i) | 0 | 0 | 181.2 ± 5.1 | 0.23 | −42.7 ± 0.5 |
| Anti-ICAM F1 | 13 | 142.8 | 160.0 ± 3.5 | 0.18 | −39.1 ± 0.8 |
| Anti-ICAM F2 | 46 | 505.5 | 179.5 ± 5.7 | 0.25 | −43.6 ± 0.1 |
| Anti-ICAM F3 | 80 | 879.1 | 196.6 ± 3.5 | 0.20 | −45.0 ± 0.1 |
| 2L 3DNA | |||||
| IgG control (ii) | 0 | 0 | 114.4 ± 3.1 | 0.34 | −38.6 ± 2.1 |
| Anti-ICAM F1 | 6 | 545.5 | 113.0 ± 4.4 | 0.35 | −36.9 ± 1.8 |
| Anti-ICAM F2 | 14 | 1272.7 | 120.4 ± 3.0 | 0.29 | −35.3 ± 0.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roki, N.; Solomon, M.; Bowers, J.; Getts, L.; Getts, R.C.; Muro, S. Tuning Design Parameters of ICAM-1-Targeted 3DNA Nanocarriers to Optimize Pulmonary Targeting Depending on Drug Type. Pharmaceutics 2022, 14, 1496. https://doi.org/10.3390/pharmaceutics14071496
Roki N, Solomon M, Bowers J, Getts L, Getts RC, Muro S. Tuning Design Parameters of ICAM-1-Targeted 3DNA Nanocarriers to Optimize Pulmonary Targeting Depending on Drug Type. Pharmaceutics. 2022; 14(7):1496. https://doi.org/10.3390/pharmaceutics14071496
Chicago/Turabian StyleRoki, Nikša, Melani Solomon, Jessica Bowers, Lori Getts, Robert C. Getts, and Silvia Muro. 2022. "Tuning Design Parameters of ICAM-1-Targeted 3DNA Nanocarriers to Optimize Pulmonary Targeting Depending on Drug Type" Pharmaceutics 14, no. 7: 1496. https://doi.org/10.3390/pharmaceutics14071496