1. Introduction
Previous treatments for schistosomiasis have consisted of a multitude of drugs, many of which have fallen out of favor in subsequent years, due to resistance, effectiveness, cost, and side effects [
1,
2]. Oxamniquine (OXA) was an efficient drug for treating
Schistosoma mansoni infections but was ineffective against the other two main schistosome species infecting humans,
S. haematobium and
S. japonicum. Because of this species specificity, OXA was the drug of choice in Brazil [
3], where only
S. mansoni is present. However, resistance has developed in the field [
4,
5,
6,
7] and was selected for use in the laboratory [
8,
9]. Praziquantel (PZQ) was developed around the same time as OXA but had the advantage of treating infections caused by all three schistosome species. PZQ has replaced OXA as a schistosomicide in all parts of the world, because of its broader spectrum and effectiveness [
10]. PZQ has few adverse side effects and is extremely cost-effective, due to an expired patent. However, PZQ is not effective against immature parasites [
11,
12,
13]. There is some concern that the emergence of PZQ-resistant worms would be inevitable due to the use of mass chemotherapy of PZQ, adding selection pressure to the parasite populations [
14]. Evidence for resistance to PZQ has already been observed in the field and selected for in the laboratory [
14,
15,
16,
17,
18,
19]. The efficacy of OXA and PZQ is comparable, although in some cases OXA is more effective against
S. mansoni when PZQ drug failure is observed [
20].
Previous studies have identified both the mechanism of OXA activity and the mechanism for OXA resistance [
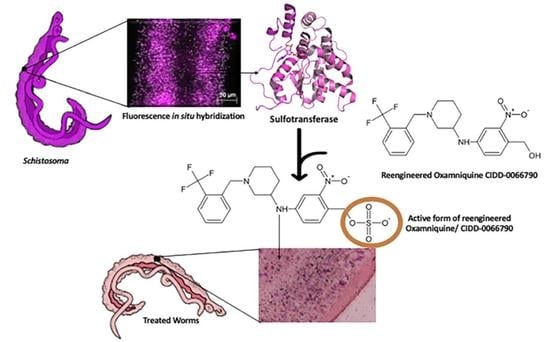
21]. OXA is a prodrug that is enzymatically activated in the parasite [
21,
22]. OXA binds to a specific
S. mansoni sulfotransferase, known as
SmSULT-OR, where it is transiently sulfated, resulting in its activation [
23]. Sulfotransferases are enzymes that catalyze the transfer of a sulfuryl group (SO
3) from a sulfate donor, such as 3′-phosphoadenosine 5′-phosphosulfate (PAPS), to substrates. Activated OXA binds to DNA and other macromolecules, resulting in the killing of the schistosome adult worms [
21,
23,
24,
25]. This affects both adult sexes but mainly males, causing the parasite to detach from the hepatoportal circulation and move into the liver where they are eliminated. Mutations in the
S. mansoni sulfotransferase (
SmSULT) are responsible for OXA resistance both in the field and in lab-derived resistant isolates [
6,
7,
21]. In a previous study, we identified an OXA derivative, CIDD-0066790, that would kill 85–100% of
S. mansoni, 40%
S. haematobium and 83%
S. japonicum [
26]. Using CIDD-0066790, in this paper, we address the question of whether the mechanism of killing for OXA derivatives that kill the three major human species of
Schistosoma is the same as for OXA. We examine the relative abundance of sulfotransferases in
S. mansoni,
S. haematobium, and
S. japonicum. In addition, we localize the distribution of
SULT within the adult male schistosome tissues. The results address questions of relative abundance of
SULTs between species, as well as identify the cells that express
SULTs and how this contributes to the killing of a multicellular eukaryotic parasite.
2. Materials and Methods
2.1. Schistosome Parasites and Animal Infections
Schistosoma mansoni,
S. haematobium and
S. japonicum were maintained in the laboratory in
Biomphalaria glabrata,
Bulinus truncatus, and
Oncomelania hupensis snails, respectively [
27]. Cercariae collected from infected snails were used to infect male Golden Syrian hamsters in accordance with an IACUC protocol (UTHSCSA IACUC Protocol #20110087AR).
2.2. Parasite Harvesting
Once the schistosomes developed into the necessary stage of worms in the mammalian host (20–90 days depending on the schistosome species and stage of development) the animals were sacrificed in accordance with IACUC protocol (UTHSCSA IACUC Protocol #20110087AR) by intraperitoneal injection using Fatal-Plus (Butler Animal Health, Dublin, OH, USA), a sodium pentobarbital solution, with 10% heparin added. Adult worms were collected by perfusion [
28]. In the case of
S. haematobium, the worms were also manually dissected out of the mesenteries and fat deposits of the bowels using forceps. The adult worms were placed in culture in media containing 1X Dulbecco’s Modified Eagle Medium (DMEM, Gibco) and 1X antibiotic/antimycotic (Ab/Am, GIBCO). Three-hour schistosomula were mechanically transformed from cercariae according to [
27].
2.3. RNA Extraction
Total RNA was obtained from frozen samples of adult S. mansoni, S. haematobium, and S. japonicum worms. All frozen samples were thawed on ice in RNAzol® RT (Molecular Research Center Inc., Cincinnati, OH, USA) and sonicated (QSonica™, Newtown, CT, USA) for 1 s 10× at 50 amps or until homogenous. RNA was extracted and purified according to manufacturer (Molecular Research Center Inc.) instructions for total RNA isolation.
2.4. cDNA Synthesis
cDNA was generated from 1 µg of total RNA using BioRad iSCRIPT cDNA Synthesis Kit according to the manufacturer’s instructions.
2.5. Quantitative Real Time PCR
Quantitative Real Time PCR (qRT-PCR) was used to determine the relative quantities of
SULT transcribed in different species of schistosomes. 150 µg of cDNA was used for relative quantification with gene-specific primers and iTaq™ Universal SYBR
® Green Supermix (BioRad, Hercules, CA, USA) containing hot-start iTaq DNA polymerase, dNTPs, MgCl2, SYBR
® Green I dye, and ROX reference dye. Primers were designed using PerlPrimer v1.21 (
Supplementary Table S1) for each
SULT gene (
SmSULT: Smp_089320;
ShSULT: Sha_104171;
SjSULT: FN317462.1) and assayed for efficiency at 1:0, 1:5, 1:25, and 1:100 cDNA concentrations.
Gapdh, β-Tubulin, and Actin were used as endogenous controls for each species. The qRT-PCR reaction was performed in 10 μL reaction and contained 5µL iTaq Universal SYBR® Green Supermix (BioRad), 3 µL cDNA (50 µg/µL), 1 µL each of forward and reverse primers 100 µM. The qRT-PCR profile was 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 60 °C for 1 min, and a final step of 60 °C for 5 min (Applied Biosystems 7500 FastReal-Time PCR System, Waltham, MA, USA).
Comparative sulfotransferase expression between schistosome species was calculated based on qRT-PCR amplification results, using an equation to adjust according to an internal reference primer and primer amplification efficiency (
Supplementary Figure S1). This approach is algebraically equivalent to the ∆∆Ct method.
2.6. Digital PCR
Digital PCR was also used to measure the absolute concentration of
SULT transcripts in total RNA. cDNA was prepared as previously described. The Bioanalytics and Single Cell Core at UTHSCSA were given cDNA at 50 ng/µL and primers at 100 µM. The sample was diluted to 20 ng/µL (“1×” concentration) and the primers were diluted to 50 µM. Digital PCR using Gene Expression Assay (Fluidigm, South San Francisco, CA, USA) was performed on a BioMark HD (Fluidigm) using EvaGreen (Biotium, Fremont, CA, USA) DNA-intercalating fluorescent dye as a reporter. The ddPCR primer sequences are in
Supplemental Table S2.
2.7. Tritiated Drug Labeling
Tritiated OXA was synthesized by the Center for Innovative Drug Discovery (CIDD) using radiolabeled NaBH4 (Moravek Biochemicals, Brea, CA, USA) at 100 mCi. For each mole of OXA in the aldehyde form, a single Ci was added and reacted at room temperature to completion and detected by thin-layer chromatography. The levels of radioactivity were determined by blotting 10 µL of the final product onto filter paper which were then counted via liquid scintillation counter (Beckman LS 6500 Scintillation Counter, Port Jefferson, NY, USA) for 10 min for each reaction.
2.8. OXA Activation Assay
The ability of a recombinant protein to activate OXA was tested by quantifying how much tritiated OXA was able to bind DNA. For each reaction, 100 μCi of [3H]OXA was solubilized in 2 µL DMSO and added to 10 µL of a 3′-phosphoadenosine-5′-phosphosulfate (PAPS) mix containing ATP and MgCl
2 at 50 mM each, and PAPS at 1 mM [
6,
29]. The radiolabeled OXA and PAPS mix was then added to 90 µL of recombinant protein with 10 ng/µL sheared
S. mansoni (gDNA) as a final target for activated [3H]OXA. The mixture was incubated at 37 °C for 2.5 h when testing worm extract. Then the reaction was stopped with 3 volumes of 1 mM sodium bicarbonate containing 0.1% SDS (
w/
v). Afterward, the reaction was extracted 3 times using 2 volumes of dichloromethane. A 10 µL aliquot of the aqueous phase was collected onto a small square of filter paper in a scintillation vial and then counted via a liquid scintillation counter (Beckman LS 6500 Scintillation Counter) for 10 min for each reaction.
2.9. RNAi
First, primers amplifying a 192–592 bp section of the gene coding region were generated using the PrimerDesign tool by IDTdna (
Supplementary Table S3). PCR was performed to produce the amplified gene section, followed by confirmation of amplification by running the PCR product on a 1% agarose gel. T7 promoters were then added to both the forward and reverse primer to flank the PCR product and confirmation of amplification was performed via 1% agarose gel. The PCR product with T7 promoters was then used as the template for in vitro transcription of the dsRNA. The dsRNA was placed in a 37 °C water bath, 4 h to overnight, then treated with DNAse to remove contaminants. Ammonium acetate (3 M) was added, followed by 100% ethanol to precipitate the RNA. The RNA was left at this step for 2 h to overnight, depending on the yield of RNA. The sample was then centrifuged at 14,000 rpm, forming an RNA pellet. The pellet was washed twice with ethanol. On the last wash, the supernatant was removed, and the ethanol was allowed to evaporate. The pellet was then resuspended in nuclease-free water. The concentration of RNA was then determined using the Thermo Scientific NanoDroprop 1000 spectrophotometer.
Adult male schistosomes were treated with 30 µg/mL dsRNA of either
S. mansoni SULT,
S. haematobium SULT, or
S. japonicum SULT or irrelevant control (
Drosophila Nautilis gene) RNAs on day 0, 3, 7 and 11. The worms were treated with 143 µM CIDD-0066790 on day 6. CIDD-0066790 is capable of killing 85–100%
S. mansoni, 40%
S. haematobium, and 83%
S. japonicum [
26]. The worms were observed for 14 days, pooled, and quick-frozen. Observation included notes on worm health, viability; lack of motility, shedding of tegument, blebbing of tegument, internal vacuolization, lethargy, and being opaque.
There were three biological replicates, 10 male worms per replicate that were pooled to conduct qPCR due to the limited number of worms. This silencing experiment was done twice to confirm gene expression ablation (pilot + full experiment). Treatment with the drug was performed once in triplicate. The qPCR samples were performed in triplicate to account for the variation of the run (technical replicates).
2.10. Whole Worm In Situ Hybridization
This protocol was adapted from Collins et al. [
30,
31].
To fix worms, freshly collected
S. mansoni,
S. haematobium, or
S. japonicum parasites were placed in a 15 mL conical tube with 10 mL DMEM + 10% FEB. 1/10th volume of a 2.5% anesthetic ethyl 3-aminobenzoate methanesulfonate (Sigma-Aldrich, St. Louis, MO, USA) was used to separate paired worms for 10 min on a rocker at room temperature. The parasites were then killed with 1 mL of 0.6 M MgCl
2 for 1 min. MgCl
2 solution was then replaced with 10 mL 4% Formaldehyde in Phosphate Buffered Saline with 0.3% Triton x-100 (PBSTx) for 4 h on a rocker at room temperature to fix the worms. After 4 h of incubation, the worms were then rinsed with 10 mL 1× PBSTx for 10 min. Worms were then dehydrated in methanol and kept at −20 °C until used. Dehydrated worms were rehydrated with 10 mL 50% methanol in 1× PBSTx for 10 min and then incubated for 10 min in 10 mL 1× PBSTx at room temperature on a rocker. Worms were bleached in a solution [9ml H
2O, 500 μL formamide, 250 μL 20× SSC (3M sodium chloride, and 3M sodium citrate, pH7.0), 400 μL 30% H
2O
2], and incubated for 1 hr at room temperature under bright light. Worms were rinsed twice with 1× PBSTx for 10 min for each wash. Worms were then incubated with 5 μg/mL Proteinase K (Invitrogen, Carlsbad, CA, USA) in 1× PBSTx for 45 min at room temperature. Worms were post-fixed in 10 mL of 4% Formaldehyde in 1× PBSTx for 10 min at room temperature. Then, worms were washed in a 1:1 ratio of PBSTx and pre-hybridization solution for 10 min at room temperature. Male parasites (5–6) were placed into 48-well-sized baskets (Intavis Bioanalytical Instruments) and incubated in pre-hybridization solution (50% deionized formamide, 5× SSC, and 1.2% H
2O
2) for 2 h at 52 °C. The worms were then incubated in 300 μL of hybridization solution (prehybridization solution with 10% dextran sulfate) and 150 ng/mL of Riboprobe generated for
SULT RNA overnight at 52 °C (
Supplemental Table S3). The worms were then washed twice, for 30 min each, in wash hybridization buffer, 2× SSC + 0.1% Triton-X, and lastly with 0.2× SSC + 0.1% Triton-X. The worms were then washed twice in 300 μL TNT (0.1M Tris.HCl (pH 7.5), 0.15M NaCl, 0.05% Tween
®-20) for 10 min each.
2.11. Oxamniquine Localization
To visualize the localization of activated OXA in
S. mansoni adult worms, [3H]OXA was incubated with living adult worms in a method similar to OXA-derivative drug screening [
32]. [3H]OXA suspended in 100% dichloromethane was allowed to evaporate and the resulting pellet weighed. The [3H]OXA pellet was solubilized in 100% DMSO to a concentration of 50 mM. [3H]OXA was administered to the adult male worms 2–24 h after harvesting from the hamsters. It was administered to the worms in media at a final concentration of 143 µM. [3H]OXA was incubated with the worms at 37 °C, 5% CO
2 for 45 min, then the media was removed without disturbing the worms. The worms were washed with plain media 3× to remove any unincorporated [3H]OXA. Worms were incubated in media for a period of 10–14 days with groups of 10 worms being removed and fixed in 4% paraformaldehyde every 2 days.
Fixed worms were embedded in paraffin, sectioned by the UTHSCSA Histology and Immunohistochemistry core facility to a thickness of 7 µm, and mounted on glass slides. The sections were de-paraffinized to water and pretreated with Photo-Flo (Kodak Professional Photo-Flo 200) for 10 min. Under dark room conditions, emulsion (Kodak NTB Emulsion) was melted in a 42 °C water bath and diluted 1:2 in distilled water. The slides were dipped into the emulsion in pairs (sample side out) and hung on a line for 4 h or until dry to the touch. The slides were packed into slide boxes containing desiccant and wrapped three times in tape and foil to prevent light exposure. The slides were incubated at 4 °C in the dark for 6 weeks. Slides were recovered from the boxes in a dark room and developed (Kodak D-19 developer) for 1 min. Slides were then washed in Millipore water for 1 min and fixed in Kodak fixative for 1 min.
Developed slides were then lightly H&E stained, mounted by the UTHSCSA Histology and Immunohistochemistry core facility, and photographed in the UTHSCSA Core Optical Imaging Facility at 100× to identify silver grain placement in the adult worms using a Nikon N-Storm Super Resolution Microscope equipped with Elipse Ti inverted TIRF microscope, and two high resolution CCD cameras: Photometrics Cool SNAP HQ2 and Andor iXon3 EMCCD.
2.13. Statistics
Statistical analysis for digital PCR studies was performed using GraphPad Prism software. One-way analysis of variance (ANOVA) was performed to compare absolute values of three individual experiments. Statistical analysis for the Kaplan-Meier curves was performed using R software scripts [
35].
4. Discussion
The three major species, that cause human schistosomiasis,
S. mansoni,
S. haematobium, and
S. japonicum, account for over 99.5% of all global cases of the disease [
36]. Currently, there is only one method of treatment, a single dose of PZQ. In 2013, the mechanism of action was elucidated for another schistosomicidal pharmaceutical, OXA. The data produced by Valentim et al. [
21] allowed for questions to be asked; why is OXA only effective at certain life stages of the parasite? Why is OXA effective in
S. mansoni but not in the other major schistosome species? Is the mode of action the same for all the OXA derivatives, that is, sulfation by a sulfotransferase, as described by Valentim et al. [
21] and Guzman et al. [
37]? Where and to what extent is the
SULT expressed in the adult parasite and does this account for the mechanism of killing? We used CIDD-0066790 for these studies as it will kill all three human species of
Schistosoma [
25].
One possible explanation for resistance to OXA by
S. haematobium and
S. japonicum, respectively, is that the activating
SULT enzyme is differentially expressed across the three species. This is a possibility given that the differences in amino acid residues in the
SULT active site across species do not seem to be responsible for differences in OXA schistosomicidal activity [
29].
Whole worm transcript levels of the sulfotransferase in
S. haematobium and
S. japonicum indicate the little-to-no presence of
SULT, which may provide a partial explanation as to why
S. haematobium and
S. japonicum are resistant to OXA [
29]. Regardless of these differences in expression, the designed OXA derivative, CIDD-0066790, could be activated by recombinant
SULT from each schistosome species (
Figure 1). As the activating agent of OXA is the sulfotransferase, and the derivative studied is based on OXA, we hypothesized the sulfotransferase is also the activating enzyme of the derivatives. To demonstrate this, the sulfotransferase was silenced in each of the schistosome species and challenged with CIDD-0066790. This approach would determine if the OXA derivative would kill the three schistosome species, indicating a change in the mode of action or if the OXA mode of action was retained. Using RNAi, silencing of the sulfotransferase in each species resulted in resistance to the OXA derivative and reinforced the notion that the mechanism of killing was the result of sulfation of CIDD-0066790 (
Figure 2,
Figure 3 and
Figure 4). Evidence to date suggests that the ability of OXA, or its derivatives, to fit into the binding pocket and accept the transfer of a sulfur group from a sulfur donor is more likely the reason for activation and subsequent killing ability [
24,
25,
29].
It was previously proposed that differences in expression was the underlying cause of OXA ineffectiveness against the other two
Schistosoma species [
21]. Therefore, we aimed to determine expression patterns between species using qPCR and highly sensitive, droplet digital PCR (ddPCR). We compared transcript levels of
SULT across the three major species using qPCR. Normally, a cross-species comparison is not ideal due to the lack of appropriate internal references. Using multiple homologous reference genes as a basis for comparison, a formula was developed to compare schistosome
SULTs across species (
Supplemental Figure S1). The initial qPCR results showed far less
SULT transcript in
S. haematobium and
S. japonicum compared
to S. mansoni (
Figure 8). This relative transcript level was independently confirmed using droplet digital PCR (ddPCR), which allowed for the absolute quantification of
SULT transcripts. Based on the ddPCR data, there is about a 13-fold difference in expression between
S. mansoni and
S. haematobium, and a 30-fold difference in expression between
S. haematobium and
S. japonicum. To stress the difference in expression between
S. mansoni and
S. japonicum, there is over a 400-fold difference between these two groups (
Figure 9). As OXA is a prodrug that requires activation via a
SULT, there is a correlation between the failure of OXA to kill the other two human schistosome species and the low amounts of
SULT in
S. haematobium and
S. japonicum. This disparity could account for OXA’s failure to kill even if the SULTs are able to bind OXA with similar affinities [
37] and activate OXA in an in vitro setting [
23].
CIDD-0066790 is effective against all three species with efficacy ranging from 40–85% depending on the schistosome species, thus the hypothesis that differences in sulfotransferase expression accounts for the difference in killing needs further exploration. While qPCR and ddPCR were informative, verification of these results is necessary at the protein level. The development of an antibody that recognizes endogenous SULT protein is a critical priority that has not yet been achieved. Polyclonal and monoclonal antibodies that recognize recombinant, but not native protein, have been produced (unpublished data). The development of an antibody that can recognize native protein would allow for further characterization of SULT protein expression in all different worm species as well as life stages.
S. mansoni is sensitive to OXA, but only in certain life stages such as the mature adult phase. Transcript levels of
SmSULT across several life stages, including cercariae, schistosomulae, immature worms (Day 20, 25, and 28), and mature adult worms correlate with OXA and OXA derivative sensitivity. Previous studies demonstrated a stage-specific susceptibility of
S. mansoni to OXA treatment [
12,
38]. Now that the mechanism of action of OXA has been elucidated [
21,
25], the expression of
SULT can be correlated with OXA killing (
Figure 5 and
Figure 6). Interestingly, at the infectious cercariae stage, the cercarial tail expresses higher levels of
SmSULT than the cercarial head. This may suggest a biological role for the sulfotransferase in the cercarial tail. Recent studies have examined the differences in transcription and translation between cercarial heads and tails [
39]. Transcriptome data is consistent with the high level of expression in cercariae [
40]. Furthermore, sex-associated differences in susceptibility to OXA treatment have been noted as adult male worms are more susceptible to treatment. Transcript level comparison between male and female adult worms also correlates with that susceptibility [
32].
To address the lingering question of where the sulfotransferase is expressed in adult worms, fluorescence in situ hybridization was used to determine expression localization. The sulfotransferase appeared to localize throughout the worm in
S. mansoni and
S. haematobium, where it appeared to localize to the parenchyma and outer tegumental region. We assume the same is true for
S. japonicum. However, the low level of
SjSULT expression was likely the reason we were not able to obtain in situ results for this species. Regarding the identification of specific cell types, the schistosome field is just starting to uncover cell-specific markers [
40]. However, transcriptome data from genome version 5 and 7 and the SchistoCyte Atlas identify
SmSULT (Smp_089320) as expressed in multiple cell types in mature and immature male and female
S. mansoni [
33,
34]. At 24 hrs post cercarial transformation to schistosomulae,
SmSULT expression begins to rise in male schistosomulae. Expression increases through days 21, and 28 and peaks at day 35. In females according to [
35,
36] expression begins to increase at day 21, peaks at day 28, and then declines, consistent with our results for male schistosomes (
Figure 5).
In conclusion, our data demonstrate that many cells in this multicellular eukaryotic pathogen express SULT, and this level of expression can promote the activation, and thereby the killing ability of OXA and its derivatives.
In order to further investigate and confirm the anatomical tissue responsible for OXA’s anthelminthic effect, we used silver grain localization to locate adduct formation with radioactive OXA. Pica-Mattoccia et al. [
24] demonstrated that OXA formed adducts with DNA, so we wanted to identify where in the worm these adducts were forming. Initial results showed a concentration of [3H]OXA along the gut lining, tegument, and subtegumental and parenchymal cells. Throughout a 12-day period, these concentrations slowly dispersed evenly throughout the tissue which correlates with the time it takes OXA (14 days) to kill adult worms in vitro. The findings presented in this article are highly significant, as they provide further insight into the mechanism of action of OXA and the OXA derivatives. As mentioned earlier, the proposed mechanism of action of these derivatives is that they are prodrugs activated by the sulfotransferase enzyme. One of two reactions is proposed to occur: (1) decay of the sulfate forms a reactive electrophilic product and subsequent alkylation of DNA, proteins, and other macromolecules leads to the death of the parasite due to the disruption of diverse cellular and metabolic functions [
21,
24], and (2) experimental evidence suggests the sulfuric acid monoester directly reacts with cellular nucleophiles in an S
N2-like reaction with sulfate as a leaving group, which disrupts cellular and metabolic functions leading to death [
23]. By visualizing sulfotransferase expression, we suggest that such widespread expression of the enzyme contributes to the killing capacity of the derivatives in a eukaryotic multicellular parasite.
The recently elucidated mechanism of action for the parent drug, OXA, has created an exploitable opening for directed drug development [
23,
25,
32]. By understanding the mechanism by which the major species of schistosomes are susceptible to OXA-derived compounds, pharmaceuticals can be designed to complement praziquantel, the drug of choice to circumvent praziquantel resistance and improve its efficacy.