Free and Open-Source Posologyr Software for Bayesian Dose Individualization: An Extensive Validation on Simulated Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Nonlinear Mixed Effects Models
- is the jth observation of subject i;
- is the number of subjects;
- is the number of observations of subject i;
- f is the function defining the structural model;
- g is the function defining the residual error model;
- is the vector of regression variables;
- for subject i, the vector is a vector of individual parameters:where,
- ○
- is a vector of fixed effects;
- ○
- is a vector of covariates;
- ○
- is a vector of normally distributed random effects, of length k, with variance-covariance matrix :
- The residual errors are normally distributed random variables centered on 0, with variance :
2.2. Estimation of Individual Parameters
2.2.1. General Strategy
2.2.2. Maximum A Posteriori
2.2.3. Markov Chain Monte Carlo
2.2.4. Sequential Importance Resampling
- Step 1 (sampling): a defined number M of parameters are sampled from a multivariate parametric proposal distribution;
- Step 2 (importance weighting): weights are computed for each of the sampled vectors, using the likelihood of the data given the parameter vector, weighted by the likelihood of the parameter vector in the proposal distribution;
- Step 3 (resampling): m parameter vectors are resampled from the M simulated vectors (M > m), with probabilities proportional to their weighting.
2.3. Implementation in Posologyr
2.4. Dosing Adjustment
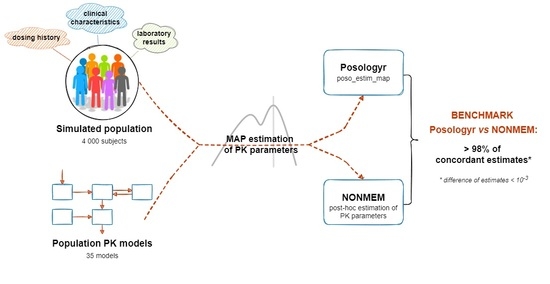
2.5. Validation
2.5.1. Point Estimate: MAP
2.5.2. Conditional Distributions
2.6. Performance Analysis
2.6.1. Point Estimate: MAP
2.6.2. Conditional Distributions
- Determination of the optimal dose to reach a concentration of 30 mg/L, 3 h after administration, with posologyr::poso_dose_conc;
- Determination of the optimal dose to achieve an AUC0-12h of 500 mg·h/L, with posologyr::poso_dose_auc;
- Determination of the time to reach a trough concentration below 0.5 mg/L after a 100 mg dose, with posologyr::poso_time_cmin.
3. Results
3.1. Point Estimate: MAP
3.2. Posterior Distribution
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B

References
- Keizer, R.J.; ter Heine, R.; Frymoyer, A.; Lesko, L.J.; Mangat, R.; Goswami, S. Model-Informed Precision Dosing at the Bedside: Scientific Challenges and Opportunities. CPT Pharmacomet. Syst. Pharmacol. 2018, 7, 785–787. [Google Scholar] [CrossRef] [PubMed]
- Frymoyer, A.; Schwenk, H.T.; Zorn, Y.; Bio, L.; Moss, J.D.; Chasmawala, B.; Faulkenberry, J.; Goswami, S.; Keizer, R.J.; Ghaskari, S. Model-Informed Precision Dosing of Vancomycin in Hospitalized Children: Implementation and Adoption at an Academic Children’s Hospital. Front. Pharmacol. 2020, 11, 551. [Google Scholar] [CrossRef]
- Proost, J.H.; Meijer, D.K.F. MW/Pharm, an Integrated Software Package for Drug Dosage Regimen Calculation and Therapeutic Drug Monitoring. Comput. Biol. Med. 1992, 22, 155–163. [Google Scholar] [CrossRef]
- Wicha, S.G.; Kees, M.G.; Solms, A.; Minichmayr, I.K.; Kratzer, A.; Kloft, C. TDMx: A Novel Web-Based Open-Access Support Tool for Optimising Antimicrobial Dosing Regimens in Clinical Routine. Int. J. Antimicrob. Agents 2015, 45, 442–444. [Google Scholar] [CrossRef] [PubMed]
- Drennan, P.; Doogue, M.; van Hal, S.J.; Chin, P. Bayesian Therapeutic Drug Monitoring Software: Past, Present and Future. Int. J. Pharmacokinet. 2018, 3, 109–114. [Google Scholar] [CrossRef]
- Maier, C.; Hartung, N.; de Wiljes, J.; Kloft, C.; Huisinga, W. Bayesian Data Assimilation to Support Informed Decision Making in Individualized Chemotherapy. CPT Pharmacomet. Syst. Pharmacol. 2020, 9, 153–164. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022. [Google Scholar]
- GNU Affero General Public License—GNU Project—Free Software Foundation. Available online: https://www.gnu.org/licenses/agpl-3.0.en.html (accessed on 15 January 2022).
- Bonate, P.L. Nonlinear Mixed Effects Models: Theory. In Pharmacokinetic-Pharmacodynamic Modeling and Simulation; Bonate, P.L., Ed.; Springer: Boston, MA, USA, 2011; pp. 233–301. ISBN 978-1-4419-9485-1. [Google Scholar]
- Kang, D.; Bae, K.-S.; Houk, B.E.; Savic, R.M.; Karlsson, M.O. Standard Error of Empirical Bayes Estimate in NONMEM® VI. Korean J. Physiol. Pharmacol. Off. J. Korean Physiol. Soc. Korean Soc. Pharmacol. 2012, 16, 97–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dosne, A.-G.; Bergstrand, M.; Karlsson, M.O. An Automated Sampling Importance Resampling Procedure for Estimating Parameter Uncertainty. J. Pharmacokinet. Pharmacodyn. 2017, 44, 509–520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Hallow, K.; James, D. A Tutorial on RxODE: Simulating Differential Equation Pharmacometric Models in R. CPT Pharmacomet. Syst. Pharmacol. 2016, 5, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Byrd, R.H.; Lu, P.; Nocedal, J.; Zhu, C. A Limited Memory Algorithm for Bound Constrained Optimization. SIAM J. Sci. Comput. 1995, 16, 1190–1208. [Google Scholar] [CrossRef]
- Comets, E.; Lavenu, A.; Lavielle, M. Parameter Estimation in Nonlinear Mixed Effect Models Using Saemix, an R Implementation of the SAEM Algorithm. J. Stat. Softw. 2017, 80, 1–41. [Google Scholar] [CrossRef] [Green Version]
- Conditional Distribution Calculation Using Monolix. Available online: https://monolix.lixoft.com/tasks/conditional-distribution/ (accessed on 15 January 2022).
- Nonmem 7.4 Users Guides. Available online: https://nonmem.iconplc.com/#/nonmem744/guides (accessed on 15 January 2022).
- Le Louedec, F.; Puisset, F.; Thomas, F.; Chatelut, É.; White-Koning, M. Easy and Reliable Maximum a Posteriori Bayesian Estimation of Pharmacokinetic Parameters with the Open-Source R Package Mapbayr. CPT Pharmacomet. Syst. Pharmacol. 2021, 10, 1208–1220. [Google Scholar] [CrossRef] [PubMed]
- Elmokadem, A.; Riggs, M.M.; Baron, K.T. Quantitative Systems Pharmacology and Physiologically-Based Pharmacokinetic Modeling With Mrgsolve: A Hands-On Tutorial. CPT Pharmacomet. Syst. Pharmacol. 2019, 8, 883–893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Using Regression Variables in Monolix. Available online: https://monolix.lixoft.com/data-and-models/regressor/ (accessed on 15 January 2022).
- Savic, R.M.; Jonker, D.M.; Kerbusch, T.; Karlsson, M.O. Implementation of a Transit Compartment Model for Describing Drug Absorption in Pharmacokinetic Studies. J. Pharmacokinet. Pharmacodyn. 2007, 34, 711–726. [Google Scholar] [CrossRef] [PubMed]




| Model Characteristics |
Model Number (Number of Estimated Parameters) | ||
|---|---|---|---|
| Oral Administration | IV Administration | ||
| Monocompartmental (default) | 1 (3) | 2 (2) | |
| Absorption | Lag time | 3 (4) | / |
| Zero-order in Central compartment | 4 (3) | / | |
| Zero-order in Depot compartment | 5 (4) | / | |
| Dual 0- and 1st orders | 6 (4) | / | |
| Dual 1st orders | 7 (4) | / | |
| Bioavailability | 8 (4) | / | |
| Distribution | Bicompartmental | 101 (4) | 102 (3) |
| Elimination | Michaelis–Menten (KM, VMAX) | 201 (4) | 202 (3) |
| Cl + Michaelis–Menten (KM) | 203 (4) | 204 (3) | |
| Cl + Michaelis–Menten (VMAX) | 205 (4) | 206 (3) | |
| Cl + Michaelis–Menten (KM, VMAX) | 207 (5) | 208 (4) | |
| Time-Varying Covariates | Time-varying Cl | 301 (3) | 302 (2) |
| Residual Error Model | Metabolite | 401 (5) | 402 (4) |
| Additive | 403 (3) | 404 (2) | |
| Mixed | 405 (3) | 406 (2) | |
| Log-additive | 407 (3) | 408 (2) | |
| Inter-individual Variability (variance) | 0.4 on all parameters | 501 (3) | / |
| 0.6 on all parameters | 502 (3) | / | |
| 0.8 on all parameters | 503 (3) | / | |
| 1 on all parameters | 504 (3) | / | |
| 2 on Cl, 0.2 on Ka, Vc | 511 (3) | / | |
| 2 on Cl, Ka, 0.2 on Vc | 512 (3) | / | |
| 2 on all parameters | 513 (3) | / | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leven, C.; Coste, A.; Mané, C. Free and Open-Source Posologyr Software for Bayesian Dose Individualization: An Extensive Validation on Simulated Data. Pharmaceutics 2022, 14, 442. https://doi.org/10.3390/pharmaceutics14020442
Leven C, Coste A, Mané C. Free and Open-Source Posologyr Software for Bayesian Dose Individualization: An Extensive Validation on Simulated Data. Pharmaceutics. 2022; 14(2):442. https://doi.org/10.3390/pharmaceutics14020442
Chicago/Turabian StyleLeven, Cyril, Anne Coste, and Camille Mané. 2022. "Free and Open-Source Posologyr Software for Bayesian Dose Individualization: An Extensive Validation on Simulated Data" Pharmaceutics 14, no. 2: 442. https://doi.org/10.3390/pharmaceutics14020442