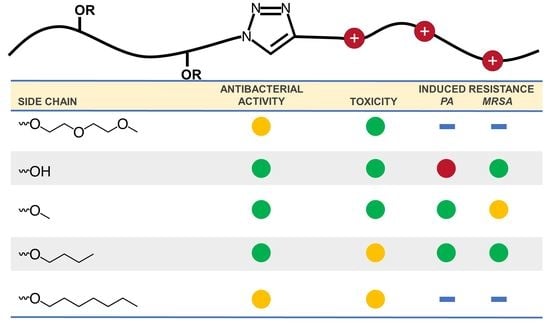
Tackling Antibiotic Resistance: Influence of Aliphatic Branches on Broad-Spectrum Antibacterial Polytriazoles against ESKAPE Group Pathogens
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Measurements
2.3. Monomer Synthesis
2.3.1. Preparation of Monomer 2
2.3.2. Preparation of Monomer 3
2.3.3. Preparation of Monomer 4
2.4. Polymer Synthesis
2.4.1. Catalytic Method
2.4.2. Thermal Method
2.4.3. Removal of the N-Boc Protecting Groups of Polytriazoles
2.5. Antibacterial Activity
2.6. Time-Kill Curves
2.7. Induced Resistance Test
2.8. Hemolysis Assay
2.9. Cytotoxicity Assay
2.10. Cytoplasmic Membrane Depolarization Assay [51,52]
2.11. Outer Membrane Permeabilization [51,52]
2.12. Inner Membrane Permeabilization [51]
2.13. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and Characterization
3.2. Gel Permeation Chromatography
3.3. Thermal Analysis
3.4. Antibacterial Activity
3.5. Bactericidal Kinetics
3.6. Induced Resistance
3.7. Membrane Stability Studies
3.8. Hemolytic Activity
3.9. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Debabov, D. Antibiotic Resistance: Origins, Mechanisms, Approaches to Counter. Appl. Biochem. Microbiol. 2013, 49, 665–671. [Google Scholar] [CrossRef]
- Si, Z.; Zheng, W.; Prananty, D.; Li, J.; Koh, C.H.; Kang, E.-T.; Pethe, K.; Chan-Park, M.B. Polymers as Advanced Antibacterial and Antibiofilm Agents for Direct and Combination Therapies. Chem. Sci. 2022, 13, 345–364. [Google Scholar] [CrossRef] [PubMed]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet Infect Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Jonas, O.B.; Irwin, A.; Berthe, F.C.J.; Le Gall, F.G.; Marquez, P.V. Drug-Resistant Infections: A Threat to Our Economic Future. Available online: https://documents.worldbank.org/en/publication/documents-reports/documentdetail/323311493396993758/final-report (accessed on 1 June 2022).
- World Health Organization. No Time to Wait: Securing the Future from Drug-Resistant Infections. Available online: https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections (accessed on 29 April 2019).
- Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available online: https://amr-review.org/Publications.html (accessed on 1 June 2022).
- Alfei, S.; Schito, A.M. Positively Charged Polymers as Promising Devices against Multidrug Resistant Gram-Negative Bacteria: A Review. Polymers 2020, 12, 1195. [Google Scholar] [CrossRef] [PubMed]
- Konai, M.M.; Bhattacharjee, B.; Ghosh, S.; Haldar, J. Recent Progress in Polymer Research to Tackle Infections and Antimicrobial Resistance. Biomacromolecules 2018, 19, 1888–1917. [Google Scholar] [CrossRef]
- Ghosh, S.; Mukherjee, S.; Patra, D.; Haldar, J. Polymeric Biomaterials for Prevention and Therapeutic Intervention of Microbial Infections. Biomacromolecules 2022, 23, 592–608. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, K.; Reghu, S.; Lin, Y.; Chan-Park, M.B.; Liu, X.-W. Synthesis of Antibacterial Glycosylated Polycaprolactones Bearing Imidazoliums with Reduced Hemolytic Activity. Biomacromolecules 2019, 20, 949–958. [Google Scholar] [CrossRef]
- Cuervo-Rodríguez, R.; Muñoz-Bonilla, A.; Araujo, J.; Echeverría, C.; Fernández-García, M. Influence of Side Chain Structure on the Thermal and Antimicrobial Properties of Cationic Methacrylic Polymers. Eur. Polym. J. 2019, 117, 86–93. [Google Scholar] [CrossRef]
- Rangel-Núñez, C.; Ramírez-Trujillo, C.; Hakkou, K.; Suárez-Cruz, A.; Molina-Pinilla, I.; Bueno-Martínez, M. Regiospecific vs. Non Regiospecific Click Azide-Alkyne Polymerization: In Vitro Study of Water-Soluble Antibacterial Poly(Amide Aminotriazole)S. Mater. Sci. Eng. C 2021, 125, 112113. [Google Scholar] [CrossRef]
- Kopiasz, R.J.; Tomaszewski, W.; Kuźmińska, A.; Chreptowicz, K.; Mierzejewska, J.; Ciach, T.; Jańczewski, D. Hydrophilic Quaternary Ammonium Ionenes—Is There an Influence of Backbone Flexibility and Topology on Antibacterial Properties? Macromol. Biosci. 2020, 20, 2000063. [Google Scholar] [CrossRef]
- Mukherjee, S.; Barman, S.; Mukherjee, R.; Haldar, J. Amphiphilic Cationic Macromolecules Highly Effective against Multi-Drug Resistant Gram-Positive Bacteria and Fungi with No Detectable Resistance. Front. Bioeng. Biotechnol. 2020, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Arora, A.; Mishra, A. Antibacterial Polymers—A Mini Review. Mater. Today Proc. 2018, 5, 17156–17161. [Google Scholar] [CrossRef]
- Liu, L.; Courtney, K.C.; Huth, S.W.; Rank, L.A.; Weisblum, B.; Chapman, E.R.; Gellman, S.H. Beyond Amphiphilic Balance: Changing Subunit Stereochemistry Alters the Pore-Forming Activity of Nylon-3 Polymers. J. Am. Chem. Soc. 2021, 143, 3219–3230. [Google Scholar] [CrossRef] [PubMed]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chem. Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Rostovtsev, V.V.; Green, L.G.; Fokin, V.V.; Sharpless, K.B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chemie Int. Ed. 2002, 41, 2596–2599. [Google Scholar] [CrossRef]
- Tornøe, C.W.; Christensen, C.; Meldal, M. Peptidotriazoles on Solid Phase: [1,2,3]-Triazoles by Regiospecific Copper(I)-Catalyzed 1,3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J. Org. Chem. 2002, 67, 3057–3064. [Google Scholar] [CrossRef]
- Lutz, J.F. 1,3-Dipolar Cycloadditions of Azides and Alkynes: A Universal Ligation Tool in Polymer and Materials Science. Angew. Chem. Int. Ed. 2007, 46, 1018–1025. [Google Scholar] [CrossRef]
- Manova, R.; Vanbeek, T.A.; Zuilhof, H. Surface Functionalization by Strain-Promoted Alkyne-Azide Click Reactions. Angew. Chem. Int. Ed. 2011, 50, 5428–5430. [Google Scholar] [CrossRef]
- Jewett, J.C.; Bertozzi, C.R. Cu-Free Click Cycloaddition Reactions in Chemical Biology. Chem. Soc. Rev. 2010, 39, 1272. [Google Scholar] [CrossRef]
- Derks, Y.H.W.; Rijpkema, M.; Amatdjais-Groenen, H.I.V.; Loeff, C.C.; de Roode, K.E.; Kip, A.; Laverman, P.; Lütje, S.; Heskamp, S.; Löwik, D.W.P.M. Strain-Promoted Azide–Alkyne Cycloaddition-Based PSMA-Targeting Ligands for Multimodal Intraoperative Tumor Detection of Prostate Cancer. Bioconjug. Chem. 2022, 33, 194–205. [Google Scholar] [CrossRef]
- Li, K.; Fong, D.; Meichsner, E.; Adronov, A. A Survey of Strain-Promoted Azide–Alkyne Cycloaddition in Polymer Chemistry. Chem. A Eur. J. 2021, 27, 5057–5073. [Google Scholar] [CrossRef] [PubMed]
- Qin, A.; Tang, L.; Lam, J.W.Y.; Jim, C.K.W.; Yu, Y.; Zhao, H.; Sun, J.Z.; Tang, B.Z. Metal-Free Click Polymerization: Synthesis and Photonic Properties of Poly(Aroyltriazole)S. Adv. Funct. Mater. 2009, 19, 1891–1900. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Sun, J.Z.; Hu, R.; Qin, A.; Tang, B.Z. Metal-Free Click Polymerization of Propiolates and Azides: Facile Synthesis of Functional Poly(Aroxycarbonyltriazole)S. Polym. Chem. 2012, 3, 1075. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Luo, Q.; Wan, X.; Petrović, Z.S.; Shah, B.K. Biopolymers from Vegetable Oils via Catalyst- and Solvent-Free “Click” Chemistry: Effects of Cross-Linking Density SuppInf. Biomacromolecules 2012, 13, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Radojčić, D.; Hairabedian, D.; Wan, X.; Petrović, Z.S. Alkynated and Azidated Octadecane as Model Compounds for Kinetic Studies of Huisgen 1,3-Dipolar Cycloaddition in Vegetable Oils. Eur. J. Lipid Sci. Technol. 2015, 117, 266–270. [Google Scholar] [CrossRef]
- Molina-Pinilla, I.; Bueno-Martínez, M.; Hakkou, K.; Galbis, J.A. Linear Poly(Amide Triazole)s Derived from D-Glucose. J. Polym. Sci. Part A Polym. Chem. 2014, 52, 629–638. [Google Scholar] [CrossRef]
- Döhler, D.; Michael, P.; Binder, W.H. CuAAC-Based Click Chemistry in Self-Healing Polymers. Acc. Chem. Res. 2017, 50, 2610–2620. [Google Scholar] [CrossRef]
- Golas, P.L.; Matyjaszewski, K. Marrying Click Chemistry with Polymerization: Expanding the Scope of Polymeric Materials. Chem. Soc. Rev. 2010, 39, 1338–1354. [Google Scholar] [CrossRef]
- Gregory, A.; Stenzel, M.H. Complex Polymer Architectures via RAFT Polymerization: From Fundamental Process to Extending the Scope Using Click Chemistry and Nature’s Building Blocks. Prog. Polym. Sci. 2012, 37, 38–105. [Google Scholar] [CrossRef]
- Buono, P.; Duval, A.; Avérous, L.; Habibi, Y. Clicking Biobased Polyphenols: A Sustainable Platform for Aromatic Polymeric Materials. ChemSusChem 2018, 11, 2472–2491. [Google Scholar] [CrossRef]
- Shi, Y.; Cao, X.; Gao, H. The Use of Azide-Alkyne Click Chemistry in Recent Syntheses and Applications of Polytriazole-Based Nanostructured Polymers. Nanoscale 2016, 8, 4864–4881. [Google Scholar] [CrossRef] [PubMed]
- Mondal, P.; Behera, P.K.; Singha, N.K. Macromolecular Engineering in Functional Polymers via ‘Click Chemistry’ Using Triazolinedione Derivatives. Prog. Polym. Sci. 2021, 113, 101343. [Google Scholar] [CrossRef]
- Tremblay-Parrado, K.K.; García-Astrain, C.; Avérous, L. Click Chemistry for the Synthesis of Biobased Polymers and Networks Derived from Vegetable Oils. Green Chem. 2021, 23, 4296–4327. [Google Scholar] [CrossRef]
- Chiloeches, A.; Funes, A.; Cuervo-Rodríguez, R.; López-Fabal, F.; Fernández-García, M.; Echeverría, C.; Muñoz-Bonilla, A. Biobased Polymers Derived from Itaconic Acid Bearing Clickable Groups with Potent Antibacterial Activity and Negligible Hemolytic Activity. Polym. Chem. 2021, 12, 3190–3200. [Google Scholar] [CrossRef]
- Ossipov, D.A.; Hilborn, J. Poly(Vinyl Alcohol)-Based Hydrogels Formed by “Click Chemistry”. Macromolecules 2006, 39, 1709–1718. [Google Scholar] [CrossRef]
- Uliniuc, A.; Popa, M.; Hamaide, T.; Dobromir, M. New Approaches in Hydrogel Synthesis—Click Chemistry: A Review. Cellul. Chem. Technol. 2012, 46, 1–11. [Google Scholar]
- Agrahari, A.K.; Bose, P.; Jaiswal, M.K.; Rajkhowa, S.; Singh, A.S.; Hotha, S.; Mishra, N.; Tiwari, V.K. Cu(I)-Catalyzed Click Chemistry in Glycoscience and Their Diverse Applications. Chem. Rev. 2021, 121, 7638–7956. [Google Scholar] [CrossRef]
- Kaur, J.; Saxena, M.; Rishi, N. An Overview of Recent Advances in Biomedical Applications of Click Chemistry. Bioconjug. Chem. 2021, 32, 1455–1471. [Google Scholar] [CrossRef]
- Kim, E.; Koo, H. Biomedical Applications of Copper-Free Click Chemistry: In Vitro, in Vivo, and Ex Vivo. Chem. Sci. 2019, 10, 7835–7851. [Google Scholar] [CrossRef] [Green Version]
- Arslan, M.; Acik, G.; Tasdelen, M.A. The Emerging Applications of Click Chemistry Reactions in the Modification of Industrial Polymers. Polym. Chem. 2019, 10, 3806–3821. [Google Scholar] [CrossRef]
- Li, H.Y.; Chen, C.Y.; Cheng, H.T.; Chu, Y.H. Exploiting 1,2,3-Triazolium Ionic Liquids for Synthesis of Tryptanthrin and Chemoselective Extraction of Copper(II) Ions and Histidine-Containing Peptides. Molecules 2016, 21, 1355. [Google Scholar] [CrossRef] [PubMed]
- Gorzynski Smith, J.; Drozda, S.E.; Petraglia, S.P.; Quinn, N.R.; Rice, E.M.; Taylor, B.S.; Viswanathan, M. Regioselective Synthesis of Allyltrimethylsilanes from Allylic Halides and Allylic Sulfonates. Application to the Synthesis of 2,3-Bis(Trimethylsilyl)Alk-l-Enes. J. Org. Chem. 1984, 49, 4112–4120. [Google Scholar] [CrossRef]
- Hakkou, K.; Bueno-Martínez, M.; Molina-Pinilla, I.; Galbis, J.A. Degradable Poly(Ester Triazole)s Based on Renewable Resources. J. Polym. Sci. Part A Polym. Chem. 2015, 53, 2481–2493. [Google Scholar] [CrossRef]
- Suárez-Cruz, A.; Molina-Pinilla, I.; Hakkou, K.; Rangel-Núñez, C.; Bueno-Martínez, M. Novel Poly(Azoamide Triazole)s Containing Twin Azobenzene Units in the Backbone. Synthesis, Characterization, and in Vitro Degradation Studies. Polym. Degrad. Stab. 2021, 193, 109726. [Google Scholar] [CrossRef]
- Srinivasachari, S.; Liu, Y.; Zhang, G.; Prevette, L.; Reineke, T.M. Trehalose Click Polymers Inhibit Nanoparticle Aggregation and Promote PDNA Delivery in Serum. J. Am. Chem. Soc. 2006, 128, 8176–8184. [Google Scholar] [CrossRef]
- Molina-Pinilla, I.; Hakkou, K.; Romero-Azogil, L.; Benito, E.; García-Martín, M.G.; Bueno-Martínez, M. Synthesis of Degradable Linear Cationic Poly(Amide Triazole)s with DNA-Condensation Capability. Eur. Polym. J. 2019, 113, 36–46. [Google Scholar] [CrossRef]
- Pollard, J.E.; Snarr, J.; Chaudhary, V.; Jennings, J.D.; Shaw, H.; Christiansen, B.; Wright, J.; Jia, W.; Bishop, R.E.; Savage, P.B. In Vitro Evaluation of the Potential for Resistance Development to Ceragenin CSA-13. J. Antimicrob. Chemother. 2012, 67, 2665–2672. [Google Scholar] [CrossRef] [Green Version]
- Barman, S.; Konai, M.M.; Samaddar, S.; Haldar, J. Amino Acid Conjugated Polymers: Antibacterial Agents Effective against Drug-Resistant Acinetobacter Baumannii with No Detectable Resistance. ACS Appl. Mater. Interfaces 2019, 11, 33559–33572. [Google Scholar] [CrossRef]
- Qian, Y.; Deng, S.; Cong, Z.; Zhang, H.; Lu, Z.; Shao, N.; Bhatti, S.A.; Zhou, C.; Cheng, J.; Gellman, S.H.; et al. Secondary Amine Pendant β-Peptide Polymers Displaying Potent Antibacterial Activity and Promising Therapeutic Potential in Treating MRSA-Induced Wound Infections and Keratitis. J. Am. Chem. Soc. 2022, 144, 1690–1699. [Google Scholar] [CrossRef]
- Kolb, H.C.; Sharpless, K.B. The Growing Impact of Click Chemistry on Drug Discovery. Drug Discov. Today 2003, 8, 1128–1137. [Google Scholar] [CrossRef]
- Agalave, S.G.; Maujan, S.R.; Pore, V.S. Click Chemistry: 1,2,3-Triazoles as Pharmacophores. Chem. Asian J. 2011, 6, 2696–2718. [Google Scholar] [CrossRef] [PubMed]
- Jing, C.; Suzuki, Y.; Matsumoto, A. Thermal Decomposition of Methacrylate Polymers Containing Tert-Butoxycarbonyl Moiety. Polym. Degrad. Stab. 2019, 166, 145–154. [Google Scholar] [CrossRef]
- Oh, J.; Kim, S.J.; Oh, M.K.; Khan, A. Antibacterial Properties of Main-Chain Cationic Polymers Prepared through Amine-Epoxy “Click” Polymerization. RSC Adv. 2020, 10, 26752–26755. [Google Scholar] [CrossRef] [PubMed]
- Chaderjian, W.B.; Chin, E.T.; Harris, R.J.; Etcheverry, T.M. Effect of Copper Sulfate on Performance of a Serum-Free CHO Cell Culture Process and the Level of Free Thiol in the Recombinant Antibody Expressed. Biotechnol. Prog. 2005, 21, 550–553. [Google Scholar] [CrossRef]
- Amsterdam, D. Antibiotics in Laboratory Medicine, 6th ed.; Wolters Kluver: Philadelphia, PA, USA, 2014; ISBN 9781451176759. [Google Scholar]
- Zhong, G.; Cheng, J.; Liang, Z.C.; Xu, L.; Lou, W.; Bao, C.; Ong, Z.Y.; Dong, H.; Yang, Y.Y.; Fan, W. Short Synthetic β-Sheet Antimicrobial Peptides for the Treatment of Multidrug-Resistant Pseudomonas Aeruginosa Burn Wound Infections. Adv. Healthc. Mater. 2017, 6, 1601134. [Google Scholar] [CrossRef]
- Olesen, S.W.; Barnett, M.L.; MacFadden, D.R.; Brownstein, J.S.; Hernández-Díaz, S.; Lipsitch, M.; Grad, Y.H. The Distribution of Antibiotic Use and Its Association with Antibiotic Resistance. Elife 2018, 7, e39435. [Google Scholar] [CrossRef]
- Zhou, W.; Du, Y.; Li, X.; Yao, C. Lipoic Acid Modified Antimicrobial Peptide with Enhanced Antimicrobial Properties. Bioorg. Med. Chem. 2020, 28, 115682. [Google Scholar] [CrossRef]
- Benarroch, J.M.; Asally, M. The Microbiologist’s Guide to Membrane Potential Dynamics. Trends Microbiol. 2020, 28, 304–314. [Google Scholar] [CrossRef] [Green Version]
- Parachin, N.S.; Franco, O.L. New Edge of Antibiotic Development: Antimicrobial Peptides and Corresponding Resistance. Front. Microbiol. 2014, 5, 905–917. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.; Fonseca, A.; Mendonça, P.; Branco, R.; Serra, A.; Morais, P.; Coelho, J. Recent Developments in Antimicrobial Polymers: A Review. Materials 2016, 9, 599. [Google Scholar] [CrossRef] [Green Version]
- King, A.; Chakrabarty, S.; Zhang, W.; Zeng, X.; Ohman, D.E.; Wood, L.F.; Abraham, S.; Rao, R.; Wynne, K.J. High Antimicrobial Effectiveness with Low Hemolytic and Cytotoxic Activity for PEG/Quaternary Copolyoxetanes. Biomacromolecules 2014, 15, 456–467. [Google Scholar] [CrossRef] [PubMed]
- Uppu, D.S.S.M.; Akkapeddi, P.; Manjunath, G.B.; Yarlagadda, V.; Hoque, J.; Haldar, J. Polymers with Tunable Side-Chain Amphiphilicity as Non-Hemolytic Antibacterial Agents. Chem. Commun. 2013, 49, 9389–9391. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Paino, M.; Muñoz-Bonilla, A.; López-Fabal, F.; Gómez-Garcés, J.L.; Heuts, J.P.A.; Fernández-García, M. Effect of Glycounits on the Antimicrobial Properties and Toxicity Behavior of Polymers Based on Quaternized DMAEMA. Biomacromolecules 2015, 16, 295–303. [Google Scholar] [CrossRef] [PubMed]











| Polymer | Yield (%) | Mw | Mw/Mn |
|---|---|---|---|
| P2C | 100 | 269,200 | 2.0 |
| P2T | 85 | 139,800 | 2.2 |
| P3T | 80 | 43,000 | 1.3 |
| P4C | 95 | 136,400 | 1.3 |
| P4T | 100 | 78,800 | 1.6 |
| P5C | 100 | 155,300 | 1.9 |
| P5T | 95 | 102,000 | 1.6 |
| Polymer | Tg 1 (°C) | T10% 2 (°C) | Tdec 3 (°C) |
|---|---|---|---|
| P1T | 80.0 | 219.0 | 235.0, 301.0 |
| P2C | 66.5 | 227.0 | 240.3, 300.0, 390.0 (s) |
| P2T | 61.2 | 222.7 | 239.4, 306.0, 390.0 (s) |
| P3T | 48.8 | 226.1 | 240.8, 312.5 |
| P4C | 38.2 | 232.1 | 244.1, 318.0, 354.0 (s) |
| P4T | 31.9 | 232.6 | 243.5, 317.6, 354.1 (s) |
| P5C | 28.6 | 227.0 | 241.0, 306.0, 380.0 (s) |
| P5T | 27.2 | 217.5 | 234.4, 304.1, 380.8 |
| P1TD | 112.4 | 258.0 | 309.0, 338.0 (s) |
| P2CD | 91.4 | 269.0 | 298.0, 358.0 |
| P2TD | 87.7 | 271.0 | 298.0, 353.0 |
| P3TD | 71.2 | 263.0 | 297.0, 371.0 |
| P4CD | 85.9 | 267.4 | 305.8, 360.9 |
| P4TD | 66.0 | 274.0 | 315.8, 359.8 |
| P5CD | 44.9 | 272.3 | 294.0, 368.0 |
| P5TD | 39.2 | 271.0 | 295.5, 367.5 |
| Polymer | Enterococcus faecium | Staphylococcus aureus | Klebsiella pneumoniae | Pseudomonas aeruginosa | Enterobacter spp. |
|---|---|---|---|---|---|
| P1TD | 16/16 | 16/16 | 32/32 | 32/32 | 16/16 |
| P2CD | 16/32 | 16/16 | 64/128 | 32/64 | 16/16 |
| P2TD | 16/32 | 16/16 | 256/512 | 128/512 | 16/16 |
| P3TD | 4/4 | 4/8 | 8/8 | 8/8 | 4/4 |
| P4CD | >1024 | >1024 | >1024 | 128/256 | 32/32 |
| P4TD | >1024 | >1024 | >1024 | >1024 | 64/128 |
| P5CD | 64/128 | 64/64 | >1024 | >1024 | 256/512 |
| P5TD | 256/256 | 128/128 | >1024 | >1024 | >1024 |
| Polymer | Enterococcus faecium | Staphylococcus aureus | Klebsiella pneumoniae | Pseudomonas aeruginosa | Enterobacter spp. |
|---|---|---|---|---|---|
| P1TD | >256 | >256 | >128 | >128 | >256 |
| P2CD | >64 | >64 | >16 | >32 | >64 |
| P2TD | 1 | 1 | <1 | <1 | 1 |
| P3TD | <1 | <1 | <1 | <1 | <1 |
| P4CD | <1 | <1 | <1 | <1 | <1 |
| P4TD | <1 | <1 | <1 | <1 | 16 |
| P5CD | >16 | >16 | -a | -a | -a |
| P5TD | >4 | >4 | - a | - a | >4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rangel-Núñez, C.; Molina-Pinilla, I.; Ramírez-Trujillo, C.; Suárez-Cruz, A.; Martínez, S.B.; Bueno-Martínez, M. Tackling Antibiotic Resistance: Influence of Aliphatic Branches on Broad-Spectrum Antibacterial Polytriazoles against ESKAPE Group Pathogens. Pharmaceutics 2022, 14, 2518. https://doi.org/10.3390/pharmaceutics14112518
Rangel-Núñez C, Molina-Pinilla I, Ramírez-Trujillo C, Suárez-Cruz A, Martínez SB, Bueno-Martínez M. Tackling Antibiotic Resistance: Influence of Aliphatic Branches on Broad-Spectrum Antibacterial Polytriazoles against ESKAPE Group Pathogens. Pharmaceutics. 2022; 14(11):2518. https://doi.org/10.3390/pharmaceutics14112518
Chicago/Turabian StyleRangel-Núñez, Cristian, Inmaculada Molina-Pinilla, Cristina Ramírez-Trujillo, Adrián Suárez-Cruz, Samuel Bernal Martínez, and Manuel Bueno-Martínez. 2022. "Tackling Antibiotic Resistance: Influence of Aliphatic Branches on Broad-Spectrum Antibacterial Polytriazoles against ESKAPE Group Pathogens" Pharmaceutics 14, no. 11: 2518. https://doi.org/10.3390/pharmaceutics14112518