Heterogeneous Structural Disturbance of Cell Membrane by Peptides with Modulated Hydrophobic Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Dynamic Giant Unilamellar Vesicle (GUV) Leakage Assay
2.3. Atomic Force Microscopy (AFM) Characterization
2.4. Molecular Dynamics (MD) Simulations
3. Results and Discussion
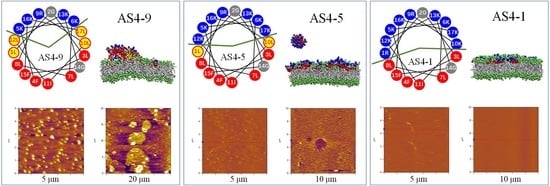
3.1. Structure and Property of AS4-1, AS4-5, and AS4-9 Peptides
3.2. Membrane Permeabilization Ability of AS4-1, AS4-5, and AS4-9
3.3. Structural Disturbance of Membranes by Peptides
3.4. Peptide-Induced Membrane Disturbance in MD Simulations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sancho-Vaello, E.; Zeth, K. Antimicrobial peptides: Has their time arrived? Future Microbiol. 2015, 10, 1103–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farsinejad, S.; Gheisary, Z.; Ebrahimi Samani, S.; Alizadeh, A.M. Mitochondrial targeted peptides for cancer therapy. Tumour Biol. 2015, 36, 5715–5725. [Google Scholar] [CrossRef] [PubMed]
- Piktel, E.; Niemirowicz, K.; Wnorowska, U.; Watek, M.; Wollny, T.; Gluszek, K.; Gozdz, S.; Levental, I.; Bucki, R. The role of cathelicidin LL-37 in cancer development. Arch. Immunol. Ther. Exp. 2016, 64, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368, 487. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Debatin, K.-M.; Poncet, D.; Kroemer, G. Chemotherapy: Targeting the mitochondrial cell death pathway. Oncogene 2002, 21, 8786–8803. [Google Scholar] [CrossRef] [Green Version]
- Radis-Baptista, G.; Campelo, I.S.; Morlighem, J.R.L.; Melo, L.M.; Freitas, V.J.F. Cell-penetrating peptides (CPPs): From delivery of nucleic acids and antigens to transduction of engineered nucleases for application in transgenesis. J. Biotechnol. 2017, 252, 15–26. [Google Scholar] [CrossRef] [Green Version]
- Mookherjee, N.; Anderson, M.A.; Haagsman, H.P.; Davidson, D.J. Antimicrobial host defence peptides: Functions and clinical potential. Nat. Rev. Drug Discov. 2020, 19, 311–332. [Google Scholar] [CrossRef]
- Greber, K.E.; Dawgul, M. Antimicrobial peptides under clinical trials. Curr. Top. Med. Chem. 2017, 17, 620–628. [Google Scholar] [CrossRef]
- Ulmschneider, J.P. Charged antimicrobial peptides can translocate across membranes without forming channel-like pores. Biophys. J. 2017, 113, 73–81. [Google Scholar] [CrossRef]
- Huang, H.W. Action of antimicrobial peptides: Two-state model. Biochemistry 2000, 39, 8347–8352. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C. How does melittin permeabilize membranes? Biophys. J. 2018, 114, 251–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Melo, M.N.; Ferre, R.; Castanho, M.A. Antimicrobial peptides: Linking partition, activity and high membrane-bound concentrations. Nat. Rev. Microbiol. 2009, 7, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Mechler, A.; Praporski, S.; Atmuri, K.; Boland, M.; Separovic, F.; Martin, L.L. Specific and selective peptide-membrane interactions revealed using quartz crystal microbalance. Biophys. J. 2007, 93, 3907–3916. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Choi, H.; Weisshaar, J.C. Melittin-induced permeabilization, re-sealing, and re-permeabilization of E. coli membranes. Biophys. J. 2018, 114, 368–379. [Google Scholar] [CrossRef] [Green Version]
- Bodescu, M.A.; Rosenkotter, F.; Fritz, J. Time lapse AFM on vesicle formation from mixed lipid bilayers induced by the membrane-active peptide melittin. Soft Matter 2017, 13, 6845–6851. [Google Scholar] [CrossRef]
- Ge, Y.; Liu, J.; Dou, Y.; Chen, Z.; Li, Z.; Yang, K.; Yuan, B.; Kang, Z. A real-time and in-situ monitoring of the molecular interactions between drug carrier polymers and a phospholipid membrane. Colloids Surf. B Biointerfaces 2022, 209, 112161. [Google Scholar] [CrossRef]
- Sun, S.; Xia, Y.; Liu, J.; Dou, Y.; Yang, K.; Yuan, B.; Kang, Z. Real-time monitoring the interfacial dynamic processes at model cell membranes: Taking cell penetrating peptide TAT as an example. J. Colloid Interface Sci. 2022, 609, 707–717. [Google Scholar] [CrossRef]
- Xu, C.; Ma, W.; Wang, K.; He, K.; Chen, Z.; Liu, J.; Yang, K.; Yuan, B. Correlation between single-molecule dynamics and biological functions of antimicrobial peptide melittin. J. Phys. Chem. Lett. 2020, 11, 4834–4841. [Google Scholar] [CrossRef]
- Ma, L.; Hu, S.; He, X.; Yang, N.; Chen, L.; Yang, C.; Ye, F.; Wei, T.; Li, M. Detection of tBid oligomerization and membrane permeabilization by graphene-based single-molecule surface-induced fluorescence attenuation. Nano Lett. 2019, 19, 6937–6944. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qian, Z.; Ma, L.; Hu, S.; Nong, D.; Xu, C.; Ye, F.; Lu, Y.; Wei, G.; Li, M. Single-molecule visualization of dynamic transitions of pore-forming peptides among multiple transmembrane positions. Nat. Commun. 2016, 7, 12906. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.-H.; Javanainen, M.; Martinez-Seara, H.; Metzler, R.; Vattulainen, I. Protein crowding in lipid bilayers gives rise to non-gaussian anomalous lateral diffusion of phospholipids and proteins. Phys. Rev. X 2016, 6, 021006. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Sun, S.; Li, W.; Zhang, Z.; Lin, Z.; Xia, Y.; Yuan, B.; Yang, K. Individual roles of peptides PGLa and Magainin 2 in synergistic membrane poration. Langmuir 2020, 36, 7190–7199. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, S.; Li, J.; Yuan, B.; Yang, K.; Ma, Y. Molecular details on the intermediate states of melittin action on a cell membrane. Biochim. Biophys. Acta Biomembr. 2018, 1860, 2234–2241. [Google Scholar] [CrossRef]
- Hong, J.; Lu, X.; Deng, Z.; Xiao, S.; Yuan, B.; Yang, K. How melittin inserts into cell membrane: Conformational changes, inter-peptide cooperation, and disturbance on the membrane. Molecules 2019, 24, 1775. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Lin, Z.; Yang, K.; Yuan, B. Single molecular kinetics during the interactions between melittin and a bi-component lipid membrane. Acta Phys. Sin. 2020, 69, 108701. [Google Scholar] [CrossRef]
- Porto, W.F.; Irazazabal, L.; Alves, E.S.F.; Ribeiro, S.M.; Matos, C.O.; Pires, A.S.; Fensterseifer, I.C.M.; Miranda, V.J.; Haney, E.F.; Humblot, V.; et al. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 2018, 9, 1490. [Google Scholar] [CrossRef] [Green Version]
- Paterson, D.J.; Tassieri, M.; Reboud, J.; Wilson, R.; Cooper, J.M. Lipid topology and electrostatic interactions underpin lytic activity of linear cationic antimicrobial peptides in membranes. Proc. Natl. Acad. Sci. USA 2017, 114, E8324–E8332. [Google Scholar] [CrossRef] [Green Version]
- Ciumac, D.; Gong, H.; Hu, X.; Lu, J.R. Membrane targeting cationic antimicrobial peptides. J. Colloid Interface Sci. 2019, 537, 163–185. [Google Scholar] [CrossRef]
- Zhang, M.H.; Ouyang, J.H.; Fu, L.; Xu, C.; Ge, Y.K.; Sun, S.Q.; Li, X.Y.; Lai, S.; Ke, H.T.; Yuan, B.; et al. Hydrophobicity determines the bacterial killing rate of α-helical antimicrobial peptides and influences the bacterial resistance development. J. Med. Chem. 2022, 65, 14701–14720. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ge, Y.; Lu, X.; Chen, Z.; Liu, J.; Zhang, M.; Yang, K.; Yuan, B. Membrane perturbation of fullerene and graphene oxide distinguished by pore-forming peptide melittin. Carbon 2021, 180, 67–76. [Google Scholar] [CrossRef]
- Lu, N.Y.; Yang, K.; Yuan, B.; Ma, Y.Q. Molecular response and cooperative behavior during the interactions of melittin with a membrane: Dissipative quartz crystal microbalance experiments and simulations. J. Phys. Chem. B 2012, 116, 9432–9438. [Google Scholar] [CrossRef] [PubMed]
- Rui, L.F.; Liu, J.J.; Li, J.L.; Weng, Y.Y.; Dou, U.J.; Yuan, B.; Yang, K.; Ma, Y.Q. Reduced graphene oxide directed self-assembly of phospholipid monolayers in liquid and gel phases. Biochim. Biophys. Acta-Biomembr. 2015, 1848, 1203–1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI force field: Coarse grained model for biomolecular simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Jong, D.H.; Singh, G.; Bennett, W.F.; Arnarez, C.; Wassenaar, T.A.; Schafer, L.V.; Periole, X.; Tieleman, D.P.; Marrink, S.J. Improved parameters for the Martini coarse-grained protein force field. J. Chem. Theory Comput. 2013, 9, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, T.A.; Ingolfsson, H.I.; Bockmann, R.A.; Tieleman, D.P.; Marrink, S.J. Computational lipidomics with insane: A versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015, 11, 2144–2155. [Google Scholar] [CrossRef]
- Pronk, S.; Pall, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; van der Spoel, D.; et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Liu, J.; Gou, L.; Li, J.; Yuan, B.; Yang, K.; Ma, Y. Designing melittin-graphene hybrid complexes for enhanced antibacterial activity. Adv. Healthc. Mater. 2019, 8, e1801521. [Google Scholar] [CrossRef]
- Chen, Y.; Cai, S.; Qiao, X.; Wu, M.; Guo, Z.; Wang, R.; Kuang, Y.Q.; Yu, H.; Wang, Y. As-CATH1-6, novel cathelicidins with potent antimicrobial and immunomodulatory properties from Alligator sinensis, play pivotal roles in host antimicrobial immune responses. Biochem. J. 2017, 474, 2861–2885. [Google Scholar] [CrossRef]
- Yin, L.M.; Edwards, M.A.; Li, J.; Yip, C.M.; Deber, C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012, 287, 7738–7745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, X.; Zhu, X.; Wang, J.; Dong, N.; Shan, A. Novel design of heptad amphiphiles to enhance cell selectivity, salt resistance, antibiofilm properties and their membrane-disruptive mechanism. J. Med. Chem. 2017, 60, 2257–2270. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Fischer, M.; Kaur, N.; Scheidt, H.A.; Mithu, V.S. Impact of lipid ratio on the permeability of mixed phosphatidylcholine/phosphatidylglycerol membranes in the presence of 1-dodecyl-3-methylimidazolium bromide ionic liquid. J. Phys. Chem. B 2022, 126, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Enoki, T.A.; Moreira-Silva, I.; Lorenzon, E.N.; Cilli, E.M.; Perez, K.R.; Riske, K.A.; Lamy, M.T. Antimicrobial peptide K(0)-W(6)-Hya1 induces stable structurally modified lipid domains in anionic membranes. Langmuir 2018, 34, 2014–2025. [Google Scholar] [CrossRef]
- Yuan, B.; Liu, J.J.; Deng, Z.X.; Wei, L.; Li, W.W.; Dou, Y.J.; Chen, Z.L.; Zhang, C.; Xia, Y.; Wang, J.; et al. A molecular architectural design that promises potent antimicrobial activity against multidrug-resistant pathogens. NPG Asia Mater. 2021, 13, 18. [Google Scholar] [CrossRef]
- Khadka, N.K.; Aryal, C.M.; Pan, J. Lipopolysaccharide-dependent membrane permeation and lipid clustering caused by cyclic lipopeptide colistin. ACS Omega 2018, 3, 17828–17834. [Google Scholar] [CrossRef]
- Muttenthaler, M.; King, G.F.; Adams, D.J.; Alewood, P.F. Trends in peptide drug discovery. Nat. Rev. Drug Discov. 2021, 20, 309–325. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dou, Y.; Chen, H.; Ge, Y.; Yang, K.; Yuan, B. Heterogeneous Structural Disturbance of Cell Membrane by Peptides with Modulated Hydrophobic Properties. Pharmaceutics 2022, 14, 2471. https://doi.org/10.3390/pharmaceutics14112471
Dou Y, Chen H, Ge Y, Yang K, Yuan B. Heterogeneous Structural Disturbance of Cell Membrane by Peptides with Modulated Hydrophobic Properties. Pharmaceutics. 2022; 14(11):2471. https://doi.org/10.3390/pharmaceutics14112471
Chicago/Turabian StyleDou, Yujiang, Haibo Chen, Yuke Ge, Kai Yang, and Bing Yuan. 2022. "Heterogeneous Structural Disturbance of Cell Membrane by Peptides with Modulated Hydrophobic Properties" Pharmaceutics 14, no. 11: 2471. https://doi.org/10.3390/pharmaceutics14112471