Arginine-Modified 3D-Printed Chromatographic Supports
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
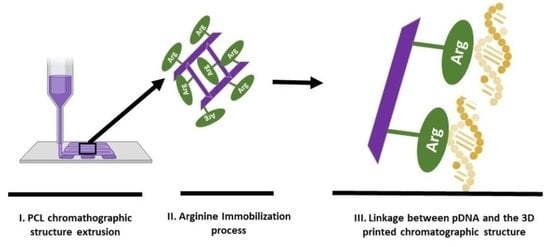
2.2.1. Production of the Chromatographic Scaffolds
2.2.2. Immobilization of Arginine in the Chromatographic Scaffolds
2.2.3. Characterization of the Arginine-Modified 3DP Chromatographic Structures
Fourier Transform Infrared Spectroscopy (FTIR)
Scanning Electron Microscopy (SEM) and Energy-Dispersive X-ray Analysis (EDX)
Micro-Computed Tomography Analysis (µ-CT)
2.2.4. Evaluation of Arginine Leakage under Different Conditions
2.2.5. Plasmid DNA Production and Isolation
The pDNA Batch Adsorption Experiments
Agarose Gel Electrophoresis
3. Results and Discussion
3.1. Chemical Evaluation of the Immobilization Process of Arginine
3.2. Morphological Evaluation of the 3D-Printed Structures
SEM
3.3. µ-CT
3.4. Assessment of the Immobilization Process Quality
3.5. The pDNA Adsorption Ability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schure, M.R.; Maier, R.S.; Kroll, D.M.; Davis, H.T. Simulation of ordered packed beds in chromatography. J. Chromatogr. A 2004, 1031, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Kalsoom, U.; Nesterenko, P.N.; Paull, B. Current and future impact of 3D printing on the separation sciences. TrAC Trends Anal. Chem. 2018, 105, 492–502. [Google Scholar] [CrossRef]
- Valente, J.F.A.; Sousa, F.; Alves, N. Additive Manufacturing Tools to Improve the Performance of Chromatographic Approaches. Trends Biotechnol. 2021, 39, 970–973. [Google Scholar] [CrossRef] [PubMed]
- Abdulhussain, N.; Nawada, S.; Currivan, S.; Passamonti, M.; Schoenmakers, P. Fabrication of polymer monoliths within the confines of non-transparent 3D-printed polymer housings. J. Chromatogr. A 2020, 1623, 461159. [Google Scholar] [CrossRef] [PubMed]
- Sousa, A.; Almeida, A.M.; Valente, J.; Queiroz, J.o.; Sousa, F. Hands-On Laboratory Class for Biopharmaceutical pDNA Quality Control. J. Chem. Educ. 2022, 99, 975–982. [Google Scholar] [CrossRef]
- Carapito, R.; Valente, J.F.A.; Queiroz, J.A.; Sousa, F. Arginine-Affinity Chromatography for Nucleic Acid (DNA and RNA) Isolation. In Affinity Chromatography; Springer: Berlin/Heidelberg, Germany, 2022; pp. 135–144. [Google Scholar]
- Fuji, T.; Anada, T.; Honda, Y.; Shiwaku, Y.; Koike, H.; Kamakura, S.; Sasaki, K.; Suzuki, O. Octacalcium phosphate–precipitated alginate scaffold for bone regeneration. Tissue Eng. Part A 2009, 15, 3525–3535. [Google Scholar] [CrossRef]
- Domingos, M.; Chiellini, F.; Cometa, S.; De Giglio, E.; Grillo-Fernandes, E.; Bártolo, P.; Chiellini, E. Evaluation of in vitro degradation of PCL scaffolds fabricated via BioExtrusion. Part 1: Influence of the degradation environment. Virtual Phys. Prototyp. 2010, 5, 65–73. [Google Scholar] [CrossRef]
- Pfaunmiller, E.L.; Paulemond, M.L.; Dupper, C.M.; Hage, D.S. Affinity monolith chromatography: A review of principles and recent analytical applications. Anal. Bioanal. Chem. 2013, 405, 2133–2145. [Google Scholar] [CrossRef] [Green Version]
- Valente, J.F.A.; Queiroz, J.A.; Sousa, F. Dilemma on plasmid DNA purification: Binding capacity vs selectivity. J. Chromatogr. A 2021, 1637, 461848. [Google Scholar] [CrossRef]
- Yew, C.H.T.; Azari, P.; Choi, J.R.; Muhamad, F.; Pingguan-Murphy, B. Electrospun polycaprolactone nanofibers as a reaction membrane for lateral flow assay. Polymers 2018, 10, 1387. [Google Scholar] [CrossRef]
- Valente, J.F.A.; Sousa, A.; Azevedo, G.; Queiroz, J.; Sousa, F. Purification of supercoiled p53-encoding plasmid using an arginine-modified macroporous support. J. Chromatogr. A 2020, 1618, 460890. [Google Scholar] [CrossRef] [PubMed]
- Pourrostam-Ravadanaq, P.; Safa, K.D.; Abbasi, H. Study of imidazole performance as pseudo-affinity ligand in the purification of IgG from bovine milk. Anal. Biochem. 2020, 597, 113693. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.R.; Dos Santos, C.; Horta, J.; Granja, P.L.; Bártolo, P.J. A new design of an electrospinning apparatus for tissue engineering applications. Int. J. Bioprinting 2017, 3, 002. [Google Scholar] [CrossRef] [PubMed]
- Zamani, Y.; Mohammadi, J.; Amoabediny, G.; Visscher, D.O.; Helder, M.N.; Zandieh-Doulabi, B.; Klein-Nulend, J. Enhanced osteogenic activity by MC3T3-E1 pre-osteoblasts on chemically surface-modified poly (ε-caprolactone) 3D-printed scaffolds compared to RGD immobilized scaffolds. Biomed. Mater. 2018, 14, 015008. [Google Scholar] [CrossRef]
- Vidič, J.; Podgornik, A.; Jančar, J.; Frankovič, V.; Košir, B.; Lendero, N.; Čuček, K.; Krajnc, M.; Štrancar, A. Chemical and chromatographic stability of methacrylate-based monolithic columns. J. Chromatogr. A 2007, 1144, 63–71. [Google Scholar] [CrossRef]
- Deshmukh, N.; Lali, A. Adsorptive purification of pDNA on superporous rigid cross-linked cellulose matrix. J. Chromatogr. B 2005, 818, 5–10. [Google Scholar] [CrossRef]
- Al-Bokari, M.; Cherrak, D.; Guiochon, G. Determination of the porosities of monolithic columns by inverse size-exclusion chromatography. J. Chromatogr. A 2002, 975, 275–284. [Google Scholar] [CrossRef]
- Yang, L.; Harding, J.D.; Ivanov, A.V.; Ramasubramanyan, N.; Dong, D.D. Effect of cleaning agents and additives on Protein A ligand degradation and chromatography performance. J. Chromatogr. A 2015, 1385, 63–68. [Google Scholar] [CrossRef]
- Linhult, M.; Gülich, S.; Gräslund, T.; Simon, A.; Karlsson, M.; Sjöberg, A.; Nord, K.; Hober, S. Improving the tolerance of a protein a analogue to repeated alkaline exposures using a bypass mutagenesis approach. Proteins Struct. Funct. Bioinform. 2004, 55, 407–416. [Google Scholar] [CrossRef]
- Hober, S.; Nord, K.; Linhult, M. Protein A chromatography for antibody purification. J. Chromatogr. B 2007, 848, 40–47. [Google Scholar] [CrossRef]
- Soares, A.; Queiroz, J.A.; Sousa, F.; Sousa, A. Purification of human papillomavirus 16 E6/E7 plasmid deoxyribonucleic acid-based vaccine using an arginine modified monolithic support. J. Chromatogr. A 2013, 1320, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, S.; de Alcântara Pessoa Filho, P.; Sousa, F.; Azzoni, A.R. Arginine and di-arginine ligands for plasmid DNA purification using negative chromatography. Sep. Purif. Technol. 2018, 202, 281–289. [Google Scholar] [CrossRef]
- Martins, R.; Queiroz, J.; Sousa, F. New approach in RNA quantification using arginine-affinity chromatography: Potential application in eukaryotic and chemically synthesized RNA. Anal. Bioanal. Chem. 2013, 405, 8849–8858. [Google Scholar] [CrossRef] [PubMed]
- Sousa, F.; Matos, T.; Prazeres, D.; Queiroz, J.A. Specific recognition of supercoiled plasmid DNA in arginine affinity chromatography. Anal. Biochem. 2008, 374, 432–434. [Google Scholar] [CrossRef] [PubMed]
- Luscombe, N.M.; Laskowski, R.A.; Thornton, J.M. Amino acid–base interactions: A three-dimensional analysis of protein–DNA interactions at an atomic level. Nucleic Acids Res. 2001, 29, 2860–2874. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Valente, J.F.A.; Carreira, T.S.; Dias, J.R.; Sousa, F.; Alves, N. Arginine-Modified 3D-Printed Chromatographic Supports. Pharmaceutics 2022, 14, 2266. https://doi.org/10.3390/pharmaceutics14112266
Valente JFA, Carreira TS, Dias JR, Sousa F, Alves N. Arginine-Modified 3D-Printed Chromatographic Supports. Pharmaceutics. 2022; 14(11):2266. https://doi.org/10.3390/pharmaceutics14112266
Chicago/Turabian StyleValente, Joana F. A., Tiago Soares Carreira, Juliana R. Dias, Fani Sousa, and Nuno Alves. 2022. "Arginine-Modified 3D-Printed Chromatographic Supports" Pharmaceutics 14, no. 11: 2266. https://doi.org/10.3390/pharmaceutics14112266