Intrinsic Aqueous Solubility: Mechanistically Transparent Data-Driven Modeling of Drug Substances
Abstract
:1. Introduction
2. Data & Methodology
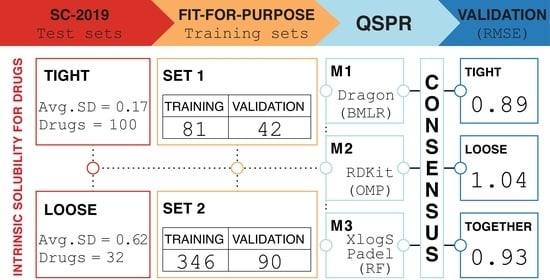
2.1. Solubility Challenge 2019 Data—The Test Sets
2.2. Intrinsic Aqueous Solubility Data for Training and Validation
2.3. Descriptor Calculation and Modeling Methods
2.4. Model Diagnostics and Applicability Domain
2.5. Availability of Models
3. Results
3.1. The Model for the Training Data Set 1
3.2. Models for Training Data Set 2
3.3. Applicability Domain and Outliers
3.4. Prediction of SC2019 Test Sets
3.5. Consensus of Models
3.6. Comparison with the Solubility Challenge 2019
4. Discussion
4.1. Fit-For-Purpose Training Set
4.2. Molecular Descriptors and Their Relevance
4.3. Selection of a Method for Model Development
4.4. Outliers in the SC2019 Test Sets
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Savjani, K.T.; Gajjar, A.K.; Savjani, J.K. Drug solubility: Importance and enhancement techniques. ISRN Pharm. 2012, 2012, 195727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taskinen, J.; Norinder, U. In Silico Predictions of Solubility. In Comprehensive Medicinal Chemistry II; Taylor, J.B., Triggle, D.J., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; Chapter 5.26; pp. 627–648. [Google Scholar] [CrossRef]
- Hörter, D.; Dressman, J.B. Influence of physicochemical properties on dissolution of drugs in the gastrointestinal tract. Adv. Drug Deliv. Rev. 2001, 46, 75–87. [Google Scholar] [CrossRef]
- Waiver of In Vivo Bioavailability and Bioequivalence Studies for Immediate-Release Solid Oral Dosage Forms Based on a Biopharmaceutics Classification System. Guidance for Industry. 2017. Available online: http://resource.nlm.nih.gov/101720038 (accessed on 27 April 2022).
- Di, L.; Kerns, E.H. Solubility Issues in Early Discovery and HTS. In Solvent Systems and Their Selection in Pharmaceutics and Biopharmaceutics; Augustijns, P., Brewster, M.E., Eds.; Biotechnology: Pharmaceutical Aspects; Springer: New York, NY, USA, 2007; Volume VI. [Google Scholar] [CrossRef]
- Pohjala, L.; Tammela, P. Aggregating behavior of phenolic compounds—A source of false bioassay results? Molecules 2012, 17, 10774–10790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birch, H.; Redman, A.D.; Letinski, D.J.; Lyon, D.Y.; Mayer, P. Determining the water solubility of difficult-to-test substances: A tutorial review. Anal. Chim. Acta 2019, 1086, 16–28. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Feeney, experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Avdeef, A.; Fuguet, E.; Llinàs, A.; Ràfols, C.; Bosch, E.; Völgyi, G.; Verbić, T.; Boldyreva, E.; Takács-Novák, K. Equilibrium solubility measurement of ionizable drugs—Consensus recommendations for improving data quality. ADMET DMPK 2016, 4, 117–178. [Google Scholar] [CrossRef] [Green Version]
- Manallack, D.T. The acid-base profile of a contemporary set of drugs: Implications for drug discovery. SAR QSAR Environ. Res. 2009, 20, 611–655. [Google Scholar] [CrossRef] [PubMed]
- Katritzky, A.R.; Maran, U.; Lobanov, V.S.; Karelson, M. Structurally Diverse QSPR Correlations of Technologically Relevant Physical Properties. J. Chem. Inf. Comput. Sci. 2000, 40, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Katritzky, A.R.; Fara, D.C.; Petrukhin, R.; Tatham, D.B.; Maran, U.; Lomaka, A.; Karelson, M. The Present Utility and Future Potential for Medicinal Chemistry of QSAR/QSPR with Whole Molecule Descriptors. Curr. Top. Med. Chem. 2002, 2, 1333–1356. [Google Scholar] [CrossRef] [Green Version]
- Karelson, M.; Diercksen, G.H.F. Models for simulationg molecular properties incondensed systems. In Problem Solving in Computational Molecular Sciences: Moleculas in Different Environments; Wilson, S., Diercksen, G.H.F., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1997; pp. 215–248. [Google Scholar]
- Karelson, M. Molecular properties and spectra in solutions. In Problem Solving in Computational Molecular Sciences: Moleculas in Different Environments; Wilson, S., Diercksen, G.H.F., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1997; pp. 353–387. [Google Scholar]
- Dearden, J.C. In Silico Prediction of Aqueous Solubility. Expert Opin. Drug Discov. 2006, 1, 31–52. [Google Scholar] [CrossRef]
- Skyner, R.E.; McDonagh, J.L.; Groom, C.R.; van Mourik, T.; Mitchell, J.B.O. A Review of Methods for the Calculation of Solution Free Energies and the Modeling of Systems in Solution. Phys. Chem. Chem. Phys. 2015, 17, 6174–6191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raevsky, O.; Grigorev, V.; Polianczyk, D.; Raevskaja, O.; Dearden, J. Aqueous Drug Solubility: What Do We Measure, Calculate and QSPR Predict? Mini-Rev. Med. Chem. 2018, 19, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Hopfinger, A.J.; Esposito, E.X.; Llinàs, A.; Glen, R.C.; Goodman, J.M. Findings of the Challenge to Predict Aqueous Solubility. J. Chem. Inf. Model. 2009, 49, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Llinàs, A.; Glen, R.C.; Goodman, J.M. Solubility Challenge: Can You Predict Solubilities of 32 Molecules Using a Database of 100 Reliable Measurements? J. Chem. Inf. Model. 2008, 48, 1289–1303. [Google Scholar] [CrossRef]
- Avdeef, A. Prediction of aqueous intrinsic solubility of druglike molecules using random forest regression trained with Wiki-pS0 database. ADMET DMPK 2020, 8, 29–77. [Google Scholar] [CrossRef] [Green Version]
- Palmer, D.S.; Mitchell, J.B.O. Is Experimental Data Quality the Limiting Factor in Predicting the Aqueous Solubility of Druglike Molecules? Mol. Pharm. 2014, 11, 2962–2972. [Google Scholar] [CrossRef] [Green Version]
- Abramov, Y.A. Major Source of Error in QSPR Prediction of Intrinsic Thermodynamic Solubility of Drugs: Solid vs. Nonsolid State Contributions? Mol. Pharm. 2015, 12, 2126–2141. [Google Scholar] [CrossRef]
- Llinas, A.; Avdeef, A. Solubility challenge revisited after ten years, with multilab shake-flask data, using thight (SD~0.17 log) and loose (SD~0.62 log) test sets. J. Chem. Inf. Model. 2019, 59, 3036–3040. [Google Scholar] [CrossRef]
- Llinas, A.; Oprisiu, I.; Avdeef, A. Findings of the Second Challenge to Predict Aqueous Solubility. J. Chem. Inf. Model. 2020, 60, 4791–4803. [Google Scholar] [CrossRef]
- Mitchell, J.B.O. Three machine learning models for the 2019 Solubility Challenge. ADMET DMPK 2020, 8, 215–250. [Google Scholar] [CrossRef]
- Lovrić, M.; Pavlović, K.; Žuvela, P.; Spataru, A.; Lučić, B.; Kern, R.; Wong, M.W. Machine learning in prediction of intrinsic aqueous solubility of drug-like compounds: Generalization, complexity, or predictive ability? J. Chemom. 2021, 35, e3349. [Google Scholar] [CrossRef]
- Falcón-Cano, G.; Molina, C.; Cabrera-Pérez, A. ADME prediction with KNIME: A retrospective contribution to the second “Solubility Challenge”. ADMET DMPK 2021, 9, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Tosca, E.M.; Bartolucci, R.; Magni, P. Application of Artificial Neural Networks to Predict the Intrinsic Solubility of Drug-Like Molecules. Pharmaceutics 2021, 13, 1101. [Google Scholar] [CrossRef]
- Francoeur, P.G.; Koes, D.R. SolTranNet—A Machine Learning Tool for Fast Aqueous Solubility Prediction. J. Chem. Inf. Model. 2021, 61, 2530–2536. [Google Scholar] [CrossRef] [PubMed]
- Falcón-Cano, G.; Molina, C.; Cabrera-Pérez, A. ADME prediction with KNIME: In silico aqueous solubility consensus model based on supervised recursive random forest approaches. ADMET DMPK 2020, 8, 251–273. [Google Scholar] [CrossRef]
- Sorkun, M.C.; Khetan, A.; Er, S. AqSolDB, a curated reference set of aqueous solubility and 2D descriptors for a diverse set of compounds. Sci. Data 2019, 6, 143. [Google Scholar] [CrossRef] [Green Version]
- Sorkun, M.C.; Khetan, A.; Er, S. AqSolDB: A curated reference set of aqueous solubility and 2D descriptors for a diverse set of compounds. Harv. Dataverse 2019. [Google Scholar] [CrossRef]
- Avdeef, A. Multi-lab intrinsic solubility measurement reproducibility in CheqSol and shake-flask methods. ADMET DMPK 2019, 7, 210–219. [Google Scholar] [CrossRef]
- Boobier, S.; Osbourn, A.; Mitchell, J.B.O. Can human experts predict solubility better than computers? J. Cheminform. 2017, 9, 63. [Google Scholar] [CrossRef]
- Bergstrom, C.A.; Wassvik, C.M.; Norinder, U.; Luthman, K.; Artursson, P. Global and local computational models for aqueous solubility prediction of druglike molecules. J. Chem. Inf. Comput. Sci. 2004, 44, 1477–1488. [Google Scholar] [CrossRef]
- Bergström, C.A.S.; Avdeef, A. Perspectives in solubility measurement and interpretation. ADMET DMPK 2019, 7, 88–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sköld, C.; Winiwarter, S.; Wernevik, J.; Bergström, F.; Engström, L.; Allen, R.; Box, K.; Comer, J.; Mole, J.; Hallberg, A.; et al. Presentation of a structurally diverse and commercially available drug data set for correlation and benchmarking studies. J. Med. Chem. 2006, 49, 6660–6671. [Google Scholar] [CrossRef] [PubMed]
- Avdeef, A.; Berger, C.M. pH-metric solubility. 3. Dissolution titration template method for solubility determination. Eur. J. Pharm. Sci. 2001, 14, 281–291. [Google Scholar] [CrossRef]
- Wassvik, C.M.; Holmen, A.G.; Bergstrom, C.A.; Zamora, I.; Artursson, P. Contribution of solid-state properties to the aqueous solubility of drugs. Eur. J. Pharm. Sci. 2006, 29, 294–319. [Google Scholar] [CrossRef]
- Baek, K.; Jeon, S.B.; Kim, B.K.; Kang, N.S. Method validation for equilibrium solubility and determination of temperature effect on the ionization constant and intrinsic solubility of drugs. J. Pharm. Sci. Emerg. Drugs 2018, 6, 1–6. [Google Scholar] [CrossRef]
- Avdeef, A.; Berger, C.M.; Brownell, C. pH-metric solubility. 2. Correlation between the acid-base titration and the saturation shake-flask solubility-pH methods. Pharm. Res. 2000, 17, 85–89. [Google Scholar] [CrossRef]
- Bergström, C.A.; Norinder, U.; Luthman, K.; Artursson, P. Experimental and computational screening models for prediction of aqueous drug solubility. Pharm. Res. 2002, 19, 182–188. [Google Scholar] [CrossRef]
- Bergström, C.A.S.; Luthman, K.; Artursson, P. Accuracy of calculated pH-dependent aqueous drug solubility. Eur. J. Pharm. Sci. 2004, 22, 387–398. [Google Scholar] [CrossRef]
- Ruusmann, V.; Maran, U. From data point timelines to a well curated data set, data mining of experimental data and chemical structure data from scientific articles, problems and possible solutions. J. Comput. Aided Mol. Des. 2013, 27, 583–603. [Google Scholar] [CrossRef]
- Standardizer, Version 19.10.0; Software for Canonicalizing Chemical Structures; Chemaxon Ltd.: Budapest, Hungary, 2019. Available online: https://www.chemaxon.com(accessed on 1 August 2019).
- Dragon, Version 6.0.40; Software for Molecular Descriptor Calculation; Talete srl: Milano, Italy, 2019. Available online: http://www.talete.mi.it/(accessed on 28 April 2022).
- Draper, N.R.; Smith, H. Applied Regression Analysis; John Wiley & Sons, Inc.: New York, NY, USA, 1966. [Google Scholar] [CrossRef]
- Karelson, M. Molecular Descriptors in QSAR/QSPR; Wiley-Interscience: New York, NY, USA, 2000; p. 448. [Google Scholar]
- CODESSA PRO, Version 1.0; University of Florida: Gainesville, FL, USA, 2001. Available online: http://www.codessa-pro.com/(accessed on 28 April 2022).
- CODESSA PRO User’s Manual; University of Florida: Gainesville, FL, USA, 2005.
- Landrum, G.A. RDKit: Open-Source Cheminformatics Software, Version 2016.03.05; RDKit: San Francisco, CA, USA. Available online: http://www.rdkit.org(accessed on 6 September 2022).
- Mallat, S.G.; Zhang, Z. Matching Pursuits with Time-Frequency Dictionaries. IEEE Trans. Signal Process. 1993, 41, 3397–3415. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Scikit-Learn: Machine Learning in Python, Version 0.18. Available online: https://scikit-learn.org/(accessed on 28 April 2022).
- Bao-Gen, D.; Yan, L.; Jie, L.; Tie-Jun, C.; Ren-Xiao, W. An Empirical Additive Model for Aqueous Solubility Computation: Success and Limitations. Acta Phys.-Chim. Sin. 2012, 28, 2249–2257. [Google Scholar] [CrossRef]
- XLOGS, Version 1.0; State Key Lab of Bioorganic Chamistry, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences: Shanghai, China. Available online: http://www.sioc-ccbg.ac.cn/?p=42&software=xlogs(accessed on 28 April 2022).
- Yap, C.W. PaDEL-Descriptor: An Open Source Software to Calculate Molecular Descriptors and Fingerprints. J. Comput. Chem. 2010, 32, 1466–1474. [Google Scholar] [CrossRef] [PubMed]
- PaDEL-Descriptor, Version 2.21. Available online: http://www.yapcwsoft.com/dd/padeldescriptor/(accessed on 28 April 2022).
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- R: A language and Environment for Statistical Computing, Version 3.5.3; R Foundation for Statistical Computing: Vienna, Austria, 2019. Available online: http://www.R-project.org/(accessed on 28 April 2022).
- Atkinson, A.C. Plots, Transformation, Regression: An Introduction to Graphical Methods of Diagnostic Regression Analysis; Oxford University Press: Oxford, UK, 1985. [Google Scholar]
- Jaworska, J.; Nikolova-Jeliazkova, N.; Aldenberg, T. QSAR applicability domain estimation by projection of the training set in descriptor space: A review. Altern. Lab. Anim. 2005, 33, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Tropsha, A.; Gramatica, P.; Gombar, V.K. The importance of being earnest: Validation is the absolute essential for successful application and interpretation of QSPR models. QSAR Comb. Sci. 2003, 22, 69–77. [Google Scholar] [CrossRef]
- Netzeva, T.I.; Worth, A.P.; Aldenberg, T.; Benigni, R.; Cronin, M.T.D.; Gramatica, P.; Jaworska, J.S.; Kahn, S.; Klopman, G.; Marchant, C.A.; et al. Current status of methods for defining the applicability domain of (quantitative) structure-activity relationships. Altern. Lab. Anim. 2005, 33, 155–173. [Google Scholar] [CrossRef]
- Ruusmann, V.; Sild, S.; Maran, U. QSAR DataBank—An approach for the digital organization and archiving of QSAR model information. J. Cheminform. 2014, 6, 25. [Google Scholar] [CrossRef]
- Ruusmann, V.; Sild, S.; Maran, U. QSAR DataBank repository: Open and linked qualitative and quantitative structure-activity relationship models. J Cheminform. 2015, 7, 32. [Google Scholar] [CrossRef] [Green Version]
- QsarDB Repository. Available online: http://qsardb.org/ (accessed on 28 April 2022).
- Oja, M.; Sild, S.; Piir, G.; Maran, U. Data for: Mechanistically transparent data-driven modeling of the intrinsic aqueous solubility of drug substances. QsarDB Repos. 2022, QDB.257. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [Green Version]
- Ghose, A.K.; Crippen, G.M. Atomic physicochemical parameters for three-dimensional-structure-directed quantitative structure-activity relationships. 2. Modeling dispersive and hydrophobic interactions. J. Chem. Inf. Comput. Sci. 1987, 27, 21–35. [Google Scholar] [CrossRef]
- Todeschini, R.; Consonni, V. Molecular Descriptors for Chemoinformatics, 2nd ed.; WILEY-VCH: Weinheim, Germany, 2009. [Google Scholar] [CrossRef]
- Estrada, E.; Ramirez, A. Edge Adjacency Relationships and Molecular Topographic Descriptors. Definition and QSAR Applications. J. Chem. Inf. Comput. Sci. 1996, 36, 837–843. [Google Scholar] [CrossRef]
- Ertl, P.; Rohde, B.; Selzer, P. Fast Calculation of Molecular Polar Surface Area as a Sum of Fragment-Based Contributions and Its Application to the Prediction of Drug Transport Properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef]
- Wildman, S.A.; Crippen, G.M. Prediction of Physicochemical Parameters by Atomic Contributions. J. Chem. Inf. Comput. Sci. 1999, 39, 868–873. [Google Scholar] [CrossRef]
- Jain, N.; Yalkowsky, S.H. Estimation of the aqueous solubility I: Application to organic nonelectrolytes. J. Pharm. Sci. 2001, 90, 234–252. [Google Scholar] [CrossRef]
- Bergström, C.A.S.; Larsson, P. Computational Prediction of Drug Solubility in Water-Based Systems: Qualitative and Quantitative Approaches Used in the Current Drug Discovery and Development Setting. Int. J. Pharm. 2018, 540, 185–193. [Google Scholar] [CrossRef]
- Avdeef, A.; Kansy, M. Can small drugs predict the intrinsic aqueous solubility of ‘beyond Rule of 5’ big drugs? ADMET DMPK 2020, 8, 180–206. [Google Scholar] [CrossRef] [PubMed]
- Ermondi, G.; Poongavanam, V.; Vallaro, M.; Kihlberg, J.; Caron, G. Solubility prediction in the bRo5 chemical space: Where are we right now? ADMET DMPK 2020, 8, 207–214. [Google Scholar] [CrossRef]
- Katritzky, A.R.; Oliferenko, A.A.; Oliferenko, P.V.; Petrukhin, R.; Tatham, D.B.; Maran, U.; Lomaka, A.; Acree, W.E. A General Treatment of Solubility. 1. The QSPR Correlation of Solvation Free Energies of Single Solutes in Series of Solvents. J. Chem. Inf. Comput. Sci. 2003, 43, 1794–1805. [Google Scholar] [CrossRef]
- Katritzky, A.R.; Oliferenko, A.A.; Oliferenko, P.V.; Petrukhin, R.; Tatham, D.B.; Maran, U.; Lomaka, A.; Acree, W.E. A General Treatment of Solubility. 2. QSPR Prediction of Free Energies of Solvation of Specified Solutes in Ranges of Solvents. J. Chem. Inf. Comput. Sci. 2003, 43, 1806–1814. [Google Scholar] [CrossRef]
- Katritzky, A.R.; Tulp, I.; Fara, D.C.; Lauria, A.; Maran, U.; Acree, W.E. A General Treatment of Solubility. 3. Principal Component Analysis (PCA) of the Solubilities of Diverse Solutes in Diverse Solvents. J. Chem. Inf. Model. 2005, 45, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Tulp, I.; Dobchev, D.A.; Katritzky, A.R.; Acree, W.; Maran, U. A General Treatment of Solubility 4. Description and Analysis of a PCA Model for Ostwald Solubility Coefficients. J. Chem. Inf. Model. 2010, 50, 1275–1283. [Google Scholar] [CrossRef]




| M1 | M2 | M3 | |
|---|---|---|---|
| Data set type | Small high-quality | Large high-variety compounds | |
| Training/validation/test | 81/42/132 | 346/90/132 | |
| Range of in training set (average, SD) | −7.1 … −1.03 (−3.55, 1.43) | −8.8 … 1.7 (−3.14, 1.64) | |
| Range of in validation set (average, SD) | −9.68 … −1.27 (−4.18, 1.77) | −10.05 … −1.24 (−4.29, 1.74) | |
| Range of in test set (average, SD) | −10.4 … −1.18 (−4.32, 1.62) | ||
| Tight test set | −6.79 … −1.18 (−4.03, 1.27) | ||
| Loose test set | −10.4 … −1.24 (−5.24, 2.18) | ||
| Descriptor calculators | Dragon | RDKit | XLOGS, PaDEL-Descriptors |
| Model development software | CODESSA Pro | scikit-learn | R statistical package |
| Descriptor selection approach | Stepwise forward selection | Orthogonal matching pursuit | Based on most common descriptors in RF models |
| Model | M1 | M2 | M3 | M_cons | |
|---|---|---|---|---|---|
| Tight test set | 0.51 | 0.52 | 0.52 | 0.57 | |
| 0.42 | 0.45 | 0.38 | 0.51 | ||
| 0.97 | 0.94 | 0.99 | 0.89 | ||
| 48% | 42% | 43% | 54% | ||
| Nr. of strong outliers | 4 | 5 | 2 | 4 | |
| Loose test set | 0.75 | 0.65 | 0.79 | 0.79 | |
| 0.74 | 0.62 | 0.75 | 0.77 | ||
| 1.1 | 1.32 | 1.06 | 1.04 | ||
| 31% | 34% | 44% | 38% | ||
| Nr. of strong outliers | 1 | 3 | 3 | 2 | |
| Tight and loose test set together | 0.66 | 0.60 | 0.65 | 0.69 | |
| 0.61 | 0.58 | 0.61 | 0.67 | ||
| 1.00 | 1.05 | 1.01 | 0.93 | ||
| 44% | 40% | 43% | 50% | ||
| Nr. of strong outliers | 5 | 8 | 5 | 6 |
| Tight Set | ||
|---|---|---|
 Cisapride Experimental: −6.78 M1: −3.92 M2: −4.64 M3: −4.07 M_cons: −4.21 |  Folic acid Experimental: −5.96 M1: −3.45 M2: −3.87 M3: −4.31 M_cons: −3.88 |  Cyclosporine A Experimental: −5.03 M1: −6.96 M2: −7.77 M3: −10.09 M_cons: −8.27 |
| Loose set | ||
 Amiodarone Experimental: −10.4 M1: −8.58 M2: −6.96 M3: −8.03 M_cons: −7.86 |  Itraconazole Experimental: −8.98 M1: −8.03 M2: −6.84 M3: −6.94 M_cons: −7.27 |  Rifabutin Experimental: −4.09 M1: −5.68 M2: −7.70 M3: −7.06 M_cons: −6.81 |
| Small Training Set (<500 comp.) | Large Training Set (>500 comp.) | |
|---|---|---|
| Many descriptors (>50) | JMSA_A (117/73/Cons. MLR) JMSA_B (117/73/extra tree reg.) JMSA_C (117/73/RFR) JHUNC_A (312/NA/ANN) | PMSA_A (2220/168/RBF) PMSA_C (7841/164, RBF) MLKC_A-C (881/NA/light GMB) RF [20] (4449/NA/RFR) |
| Fewer descriptors (<50) | M1 (UMUT_A, 81/2/MLR) M2 (UMUT_B, 346/2/MLR) M3 (UMUT_C, 346/3/MLR) M_cons (346 + 81, 7, consensus) | MCSMD (2666/7/ANN) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oja, M.; Sild, S.; Piir, G.; Maran, U. Intrinsic Aqueous Solubility: Mechanistically Transparent Data-Driven Modeling of Drug Substances. Pharmaceutics 2022, 14, 2248. https://doi.org/10.3390/pharmaceutics14102248
Oja M, Sild S, Piir G, Maran U. Intrinsic Aqueous Solubility: Mechanistically Transparent Data-Driven Modeling of Drug Substances. Pharmaceutics. 2022; 14(10):2248. https://doi.org/10.3390/pharmaceutics14102248
Chicago/Turabian StyleOja, Mare, Sulev Sild, Geven Piir, and Uko Maran. 2022. "Intrinsic Aqueous Solubility: Mechanistically Transparent Data-Driven Modeling of Drug Substances" Pharmaceutics 14, no. 10: 2248. https://doi.org/10.3390/pharmaceutics14102248