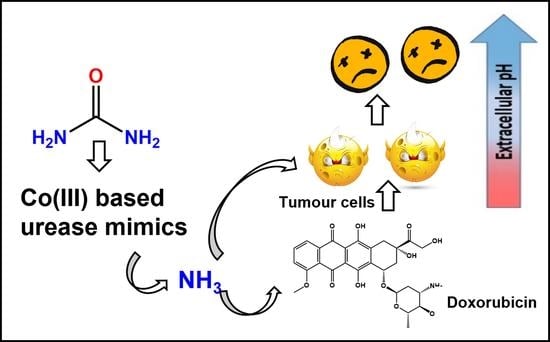
Anticancer Activity of Urease Mimetic Cobalt (III) Complexes on A549-Lung Cancer Cells: Targeting the Acidic Microenvironment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials-Physical Measurements
2.2. Cell Culture and Treatment
2.3. Stability of Co (III) Complexes
2.4. Urease Activity
2.5. Cellular Morphology-Inverted Microscopy
2.6. Cellular Proliferation-Adenosine Triphosphate (ATP) Luminescent Assay
2.7. Cytotoxicity-Lactate Dehydrogenase (LDH) Assay
2.8. Caspase 3/7 Activity
2.9. Statistical Analysis
3. Results
3.1. Synthesis, Characterization, Stability and Urease Mimetic Activity of the Complexes
3.2. Cytotoxicity-Lactate Dehydrogenase (LDH) Assay
3.3. Adenosine Triphosphate (ATP) Cell Metabolism Assay
3.4. Cellular Morphology
3.5. Trypan Blue Viability Assay
3.6. Caspase 3/7 Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sritharan, S.; Sivalingam, N. A comprehensive review on time-tested anticancer drug doxorubicin. Life Sci. 2021, 278, 119527. [Google Scholar] [CrossRef] [PubMed]
- Simões, R.; Silva, L.M.; De Oliveira, A.N.; Alves, M.T.; Pestana, R.M.C.; De Souza, I.D.P.; Oliveira, H.H.M.; Soares, C.E.; Sabino, A.D.P.; Gomes, K.B. Identification of Clinical and Laboratory Variables Associated with Cardiotoxicity Events Due to Doxorubicin in Breast Cancer Patients: A 1-Year Follow-Up Study. Cardiovasc. Toxicol. 2012, 21, 106–114. [Google Scholar] [CrossRef]
- Tripathi, R.; Kumar, A. Application of Nanorobotics for Cancer Treatment. Mater. Today Proc. 2018, 5, 9114–9117. [Google Scholar] [CrossRef]
- Hortelão, A.C.; Patiño, T.; Perez-jiménez, A.; Blanco, À.; Sánchez, S. Enzyme-Powered Nanobots Enhance Anticancer Drug Delivery. Adv. Funct. Mater. 2018, 28, 1705086. [Google Scholar] [CrossRef]
- Li, Z.; Tan, S.; Li, S.; Shen, Q.; Wang, K. Cancer drug delivery in the nano era: An overview and perspectives (Review). Oncol. Rep. 2017, 38, 611–624. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, J.; Xiong, Q.; Hornburg, D.; Tao, W.; Farokhzad, O.C. Nano–Bio Interactions in Cancer: From Therapeutics Delivery to Early Detection. Acc. Chem. Res. 2021, 54, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Farkona, S.; Diamandis, E.P.; Blasutig, I.M. Cancer immunotherapy: The beginning of the end of cancer? BMC Med. 2016, 14, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkwood, J.M.; Butterfield, L.H.; Tarhini, A.A.; Zarour, H.; Kalinski, P.; Ferrone, S. Immunotherapy of cancer in 2012. CA Cancer J. Clin. 2012, 62, 309–335. [Google Scholar] [CrossRef]
- Cao, R.; Song, W.; Ye, C.; Liu, X.; Li, L.; Li, Y.; Yao, H.; Zhou, X.; Li, L.; Shao, R. Internal enhancement of DNA damage by a novel bispecific antibody-drug conjugate-like therapeutics via blockage of mTOR and PD-L1 signal pathways in pancreatic cancer. Cancer Med. 2019, 8, 643–655. [Google Scholar] [CrossRef]
- Thomas, A.; Teicher, B.A.; Hassan, R. Antibody–drug conjugates for cancer therapy. Lancet Oncol. 2016, 17, e254–e262. [Google Scholar] [CrossRef]
- Chau, C.H.; Steeg, P.S.; Figg, W.D. Antibody–drug conjugates for cancer. Lancet 2019, 394, 793–804. [Google Scholar] [CrossRef]
- Chao, H. DOS47–Killing Cancer by Altering the Tumor Microenvironment. Drug Deliv. Technol. 2011, 11, 68–72. [Google Scholar]
- Damgaci, S.; Ibrahim-Hashim, A.; Enriquez-Navas, P.M.; Pilon-Thomas, S.; Guvenis, A.; Gillies, R.J. Hypoxia and acidosis: Immune suppressors and therapeutic targets. Immunology 2018, 154, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Wong, W.Y.; Uger, M.D.; Wisniewski, P.; Chao, H.; Tian, B. Development and Characterization of a Camelid Single Domain Antibody–Urease Conjugate That Targets Vascular Endothelial Growth Factor Receptor 2. Front. Immunol. 2017, 8, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Albini, A.; Sporn, M.B. The tumour microenvironment as a target for chemoprevention. Nat. Rev. Cancer 2007, 7, 139–147. [Google Scholar] [CrossRef]
- Dai, Y.; Xu, C.; Sun, X.; Chen, X. Nanoparticle design strategies for enhanced anticancer therapy by exploiting the tumour microenvironment. Chem. Soc. Rev. 2017, 46, 3830–3852. [Google Scholar] [CrossRef]
- Cassim, S.; Pouyssegur, J. Tumor Microenvironment: A Metabolic Player that Shapes the Immune Response. Int. J. Mol. Sci. 2019, 21, 157. [Google Scholar] [CrossRef] [Green Version]
- Petrova, V.; Annicchiarico-Petruzzelli, M.; Melino, G.; Amelio, I. The hypoxic tumour microenvironment Hypoxia and hypoxia-inducible factors. Oncogenesis 2018, 7, 10. [Google Scholar] [CrossRef]
- Wong, W.Y.; DeLuca, C.I.; Tian, B.; Wilson, I.; Molund, S.; Warriar, N.; Govindan, M.V.; Segal, D.; Chao, H. Urease-induced alkalinization of extracellular pH and its antitumor activity in human breast and lung cancers. J. Exp. Ther. Oncol. 2005, 5, 93–99. [Google Scholar]
- Pillai, S.R.; Damaghi, M.; Marunaka, Y.; Spugnini, E.P.; Fais, S.; Gillies, R.J. Causes, consequences, and therapy of tumors acidosis. Cancer Metastasis Rev. 2019, 38, 205–222. [Google Scholar] [CrossRef]
- Wojtkowiak, J.W.; Verduzco, D.; Schramm, K.J.; Gillies, R.J. Drug resistance and cellular adaptation to tumor acidic pH microenvironment. Mol. Pharm. 2011, 8, 2032–2038. [Google Scholar] [CrossRef]
- Tian, B.; Wong, W.Y.; Hegmann, E.; Gaspar, K.; Kumar, P.; Chao, H. Production and Characterization of a Camelid Single Domain Antibody-Urease Enzyme Conjugate for the Treatment of Cancer. Bioconjug. Chem. 2015, 26, 1144–1155. [Google Scholar] [CrossRef]
- Kafarski, P.; Talma, M. Recent advances in design of new urease inhibitors: A review. J. Adv. Res. 2018, 13, 101–112. [Google Scholar] [CrossRef]
- Follmer, C.J. Ureases as a target for the treatment of gastric and urinary infections. J. Clin. Pathol. 2010, 63, 424–430. [Google Scholar] [CrossRef]
- Casali, L.; Mazzei, L.; Shemchuk, O.; Honer, K.; Grepioni, F.; Ciurli, S.; Braga, D.; Baltrusaitis, J. Smart urea ionic co-crystals with enhanced urease inhibition activity for improved nitrogen cycle management. Chem. Commun. 2018, 54, 7637–7640. [Google Scholar] [CrossRef]
- Musiani, F.; Arnofi, E.; Casadio, R.; Ciurli, S. Structure-based computational study of the catalytic and inhibition mechanisms of urease. J. Biol. Inorg. Chem. 2001, 6, 300–314. [Google Scholar] [CrossRef]
- You, Z.L.; Zhang, L.; Shi, D.H.; Wang, X.L.; Li, X.F.; Ma, Y.P. Synthesis, crystal structures and urease inhibitory activity of copper(II) complexes with Schiff bases. Inorg. Chem. Commun. 2010, 13, 996–998. [Google Scholar] [CrossRef]
- Nile, S.H.; Keum, Y.S.; Nile, A.S.; Jalde, S.S.; Patel, R.V. Antioxidant, anti-inflammatory, and enzyme inhibitory activity of natural plant flavonoids and their synthesized derivatives. J. Biochem. Mol. Toxicol. 2018, 32. [Google Scholar] [CrossRef]
- Xiao, Z.P.; Wang, X.D.; Peng, Z.Y.; Huang, S.; Yang, P.; Li, Q.S.; Zhou, L.H.; Hu, X.J.; Wu, L.J.; Zhou, Y.; et al. Molecular Docking, kinetics study, and structure-activity relationship analysis of quercetin and its analogous as helicobacter pylori urease inhibitors. J. Agric. Food Chem. 2012, 60, 10572–10577. [Google Scholar] [CrossRef]
- Habala, L.; Devínsky, F.; Egger, A.E. REVIEW: Metal complexes as urease inhibitors. J. Coord. Chem. 2018, 71, 907–940. [Google Scholar] [CrossRef]
- You, Z.-L.; Ni, L.-L.; Shi, D.-H.; Bai, S. Synthesis, structures, and urease inhibitory activities of three copper(II) and zinc(II) complexes with 2-{[2-(2-hydroxyethylamino)ethylimino]methyl}-4-nitrophenol. Eur. J. Med. Chem. 2010, 45, 3196–3199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Mulrooney, S.B.; Leung, A.F.K.; Zeng, Y.; Ko, B.B.C.; Hausinger, R.P.; Sun, H. Inhibition of urease by bismuth(III): Implications for the mechanism of action of bismuth drugs. BioMetals 2006, 19, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Uprety, B.; Arderne, C.; Bernal, I. Catalytic Cleavage of the Amide Bond in Urea Using a Cobalt(III) Amino-Based Complex. Eur. J. Inorg. Chem. 2018, 2018, 5058–5067. [Google Scholar] [CrossRef]
- Saha, M.K.; Mukhopadhyay, U.; Bernal, I. Cleavage of the peptide bond of β-alanyl-l-histidine(carnosine) induced by a Co III–amine complexes: Reaction, structure and mechanism. Dalt. Trans. 2004, 1466–1473. [Google Scholar] [CrossRef]
- Saha, M.K.; Bernal, I. An unprecedented trans-oriented product from the cleavage of a dipeptide. Chem. Commun. 2003, 5, 612–613. [Google Scholar] [CrossRef]
- Rajendran, R.; Pandi, A.; Ramchary, A.; Thiagarajan, H.; Panneerselvam, J.; Niraikulam, A.; Kuppuswami, G.M.; Ramudu, K.N. Extracellular urease from Arthrobacter creatinolyticus MTCC 5604: Scale up, purification and its cytotoxic effect thereof. Mol. Biol. Rep. 2018, 46, 133–141. [Google Scholar] [CrossRef]
- Chao, H.; Wong, W.Y.; Tian, B.; Gaspar, K.J.; Kumar, P. Antibody-Urease Conjugates for Therapeutic Purposes. U.S. Patent No. 10316311, 28 July 2016. [Google Scholar]
- Chao, H. Use of Antibody-Urease Conjugates for Diagnostic and Therapeutic Purposes. International Patent No. WO2014165985A1, 16 October 2014. [Google Scholar]
- Rodryg, R.; Dariusz, K.; Cezary, S.; Aleksandra, S.; Elzbieta, W.; Steve, D.; Chao, H.; Roszkowski-Sliz, K. Phase 1 Study of Ramucirumab or Necitumumab in Combination with Osimertinib (AZD9291) in Advanced Topic: Phase I Trials Phase I/II Dose Escalation Study of L-DOS47 as a Monotherapy in Non-Squamous Non-Small Cell Lung Cancer Patients A Phase 1/2 Tri. J. Thoraic Oncol. 2017, 12, 1071–1072. [Google Scholar] [CrossRef] [Green Version]
- Cantarella, H.; Otto, R.; Soares, J.R.; de Brito Silva, A.G. Agronomic efficiency of NBPT as a urease inhibitor: A review. J. Adv. Res. 2018, 13, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, M.; Tenuta, M.; Ma, Z.; Gui, D.; Li, X.; Zeng, F.; Gao, X. Agronomic evaluation of polymer-coated urea and urease and nitrification inhibitors for cotton production under drip-fertigation in a dry climate. Sci. Rep. 2020, 10, 1472. [Google Scholar] [CrossRef] [Green Version]
- Menteşe, E.; Akyüz, G.; Emirik, M.; Baltaş, N. Synthesis, in vitro urease inhibition and molecular docking studies of some novel quinazolin-4(3H)-one derivatives containing triazole, thiadiazole and thiosemicarbazide functionalities. Bioorg. Chem. 2019, 83, 289–296. [Google Scholar] [CrossRef]
- Qu, D.; Niu, F.; Zhao, X.; Yan, K.X.; Ye, Y.T.; Wang, J.; Zhang, M.; You, Z. Synthesis, crystal structures, and urease inhibition of an acetohydroxamate-coordinated oxovanadium(V) complex derived from N???-(3-bromo-2-hydroxybenzylidene)-4-methoxybenzohydrazide. Bioorganic Med. Chem. 2015, 23, 1944–1949. [Google Scholar] [CrossRef]
- Allawi, M.H.; Abed, M.Q. A Study of Anticancer Activity for Partial Purified Urease Isolated from Lagonychium farctum Plant. Al-Nahrain J. Sci. 2018, 1, 95–99. [Google Scholar] [CrossRef] [Green Version]
- Buckingham, D.A.; Cresswell, P.J.; Sargeson, A.M. Site of Deprotonation in the Base Hydrolysis of Chloropentaaminecobalt(III) Ions. Inorg. Chem. 1975, 14, 1485–1490. [Google Scholar] [CrossRef]
- Buckingham, D.A.; Collman, J.P. Reaction of Hydroxoaquobis(Ethylenediamine)Cobalt(3) Ion With Amino Acids and Dipeptides and Their Esters and Amides. Inorg. Chem. 1967, 6, 1803–1807. [Google Scholar] [CrossRef]
- Guzei, I.A.; Arderne, C. Polymorphism of di-nitro-[tris-(2-amino-eth-yl)amine]-cobalt(III) chloride. Acta Crystallogr. Sect. C Struct. Chem. 2015, 71, 695–700. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Shen, Y.; Yang, S.; Zhang, Y. Conformational Design Principles in Total Synthesis. Angew. Chem. Int. Ed. 2020, 59, 14198–14210. [Google Scholar] [CrossRef]
- Alastair, G.; Lawson, M.D.; MacCoss, M.; Heer, J.P. Importance of Rigidity in Designing Small Molecule Drugs To Tackle Protein−Protein Interactions (PPIs) through Stabilization of Desired Conformers: Miniperspective. J. Med. Chem. 2017, 61, 4283–4289. [Google Scholar] [CrossRef]
- Arderne, C.; Batchelor, K.F.; Uprety, B.; Chandran, R.; Abrahamse, H. Reactivity trends of cobalt(III) complexes towards various amino acids based on the properties of the amino acid alkyl chains. Acta Crystallogr. Sect. C Struct. Chem. 2020, 76, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Pilot, C.; Marunaka, Y.; Fais, S. Targeting acidity in cancer and diabetes. Biochim. Biophys. Acta-Rev. Cancer 2019, 1871, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, R.; Sasaki, K.; Yoshida, K. Identification of epigallocatechin-3-gallate in green tea polyphenols as a potent inducer of p53-dependent apoptosis in the human lung cancer cell line A549. Toxicol. In Vitro 2009, 23, 834–839. [Google Scholar] [CrossRef]
- Peña, B.; Saha, S.; Barhoumi, R.; Burghardt, R.C.; Dunbar, K.R. Ruthenium(II)-Polypyridyl Compounds with π-Extended Nitrogen Donor Ligands Induce Apoptosis in Human Lung Adenocarcinoma (A549) Cells by Triggering Caspase-3/7 Pathway. Inorg. Chem. 2018, 57, 12777–12786. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, C.; Wang, Y.; Xu, L.; He, X.; Zeng, Q.; Zeng, C.; Mai, F.; Hu, B.; Ouyang, D. Chemotherapeutic paclitaxel and cisplatin differentially induce pyroptosis in A549 lung cancer cells via caspase-3/GSDME activation. Apoptosis 2019, 24, 312–325. [Google Scholar] [CrossRef]
- D’Souza, G.G.; Wagle, M.A.; Saxena, V.; Shah, A. Approaches for targeting mitochondria in cancer therapy. Biochim. Biophys. Acta 2011, 1807, 689–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plitzko, B.; Loesgen, S. Measurement of Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR) in Culture Cells for Assessment of the Energy Metabolism. Bio. Protoc. 2018, 20, 2850. [Google Scholar] [CrossRef] [PubMed]
- Vucetic, M.; Daher, B.; Cassim, S.; Meira, W.; Pouyssegur, J. Together we stand, apart we fall: How cell-to-cell contact/interplay provides resistance to ferroptosis. Cell Death Dis. 2020, 11, 789. [Google Scholar] [CrossRef] [PubMed]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uprety, B.; Chandran, R.; Arderne, C.; Abrahamse, H. Anticancer Activity of Urease Mimetic Cobalt (III) Complexes on A549-Lung Cancer Cells: Targeting the Acidic Microenvironment. Pharmaceutics 2022, 14, 211. https://doi.org/10.3390/pharmaceutics14010211
Uprety B, Chandran R, Arderne C, Abrahamse H. Anticancer Activity of Urease Mimetic Cobalt (III) Complexes on A549-Lung Cancer Cells: Targeting the Acidic Microenvironment. Pharmaceutics. 2022; 14(1):211. https://doi.org/10.3390/pharmaceutics14010211
Chicago/Turabian StyleUprety, Bhawna, Rahul Chandran, Charmaine Arderne, and Heidi Abrahamse. 2022. "Anticancer Activity of Urease Mimetic Cobalt (III) Complexes on A549-Lung Cancer Cells: Targeting the Acidic Microenvironment" Pharmaceutics 14, no. 1: 211. https://doi.org/10.3390/pharmaceutics14010211