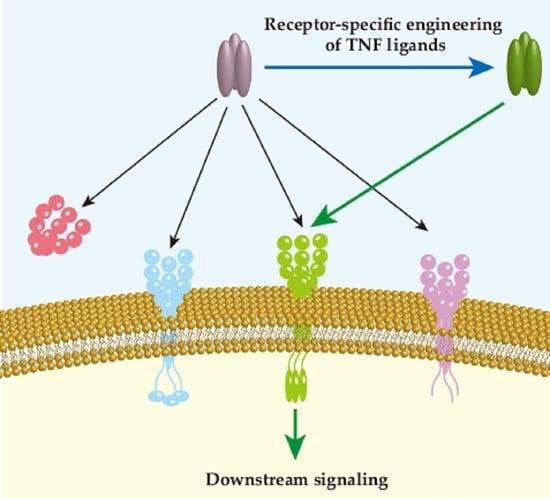
Receptor Specificity Engineering of TNF Superfamily Ligands
Abstract
:1. Introduction
2. Tumor Necrosis Factor and Lymphotoxin
2.1. Tumor Necrosis Factor and Lymphotoxin
2.2. The TNFR1 and TNFR2 Induced Signal Transduction
2.3. Engineering TNFR1-Specific Ligands
2.4. TNFR2-Specific Applications
3. TRAIL
3.1. TRAIL Induced Signaling Conduction
3.2. TRAIL Variants with Enhanced Binding to Both DR4 and DR5
3.3. TRAIL Variants with Specific Binding to DR4 or DR5
4. RANKL
4.1. RANKL Induced Signaling Conduction
4.2. RANKL Variants with Lower Binding to RANK
4.3. RANKL Variant with Lower Binding to OPG and Targeted to RANK
4.4. RANKL-Related Peptides and Antibodies That Inhibit RANK Funtion
5. FASL
6. LIGHT
6.1. LIGHT and Its Receptors
6.2. Specific Targeting HVEM and LTβR Strategy
7. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Aggarwal, B.B. Signalling pathways of the TNF superfamily: A double-edged sword. Nat. Rev. Immunol. 2003, 3, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Dostert, C.; Grusdat, M.; Letellier, E.; Brenner, D. The TNF Family of Ligands and Receptors: Communication Modules in the Immune System and Beyond. Physiol. Rev. 2019, 99, 115–160. [Google Scholar] [CrossRef] [PubMed]
- Croft, M.; Siegel, R.M. Beyond TNF: TNF superfamily cytokines as targets for the treatment of rheumatic diseases. Nat. Rev. Rheumatol. 2017, 13, 217–233. [Google Scholar] [CrossRef] [Green Version]
- Anti-TNFα Use During Elective Foot and Ankle Surgery in Patients With Rheumatoid Arthritis. Available online: https://clinicaltrials.gov/ct2/show/NCT02242474?term=TNF-α&draw=3&rank=11 (accessed on 5 January 2022).
- Targeted Stem Cells Expressing TRAIL as a Therapy for Lung Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT03298763?term=TRAIL&draw=2&rank=7 (accessed on 5 January 2022).
- Safety, Tolerability and PK/PD of JMT103 in Patients With Bone Metastases From Tumors. Available online: https://clinicaltrials.gov/ct2/show/NCT03550508?term=RANKL&draw=2&rank=13 (accessed on 5 January 2022).
- Determination of the RANKL/Osteoprotegerin Ratio in Patients With Systemic Lupus Erythematosus. Role in Osteoporosis and Cardiovascular Calcification. Available online: https://clinicaltrials.gov/ct2/show/NCT02799173?term=RANKL&draw=2&rank=4 (accessed on 5 January 2022).
- Buhrmann, C.; Shayan, P.; Aggarwal, B.B.; Shakibaei, M. Evidence that TNF-β (lymphotoxin α) can activate the inflammatory environment in human chondrocytes. Arthritis Res. Ther. 2013, 15, R202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carswell, E.A.; Old, L.J.; Kassel, R.L.; Green, S.; Fiore, N.; Williamson, B. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl. Acad. Sci. USA 1975, 72, 3666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wakeham, J.; Harkness, R.; Xing, Z. Macrophages are a significant source of type 1 cytokines during mycobacterial infection. J. Clin. Investig. 1999, 103, 1023–1029. [Google Scholar] [CrossRef] [Green Version]
- Maskos, K.; Fernandez-Catalan, C.; Huber, R.; Bourenkov, G.P.; Bartunik, H.; Ellestad, G.A.; Reddy, P.; Wolfson, M.F.; Rauch, C.T.; Castner, B.J.; et al. Crystal structure of the catalytic domain of human tumor necrosis factor-α-converting enzyme. Proc. Natl. Acad. Sci. USA 1998, 95, 3408–3412. [Google Scholar] [CrossRef] [Green Version]
- Dittrich, G.M.; Heineke, J. TNF-α signaling: TACE inhibition to put out the burning heart. PLoS Biol. 2020, 18, e3001037. [Google Scholar] [CrossRef]
- Grell, M.; Wajant, H.; Zimmermann, G.; Scheurich, P. The type 1 receptor (CD120a) is the high-affinity receptor for soluble tumor necrosis factor. Proc. Natl. Acad. Sci. USA 1998, 95, 570–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruddle, N.H.; Waksman, B.H. Cytotoxicity mediated by soluble antigen and lymphocytes in delayed hypersensitivity. 3. Analysis of mechanism. J. Exp. Med. 1968, 128, 1267–1279. [Google Scholar] [CrossRef] [Green Version]
- Kolb, W.P.; Granger, G.A. Lymphocyte in vitro cytotoxicity: Characterization of human lymphotoxin. Proc. Natl. Acad. Sci. USA 1968, 61, 1250–1255. [Google Scholar] [CrossRef] [Green Version]
- Shalaby, M.R.; Aggarwal, B.B.; Rinderknecht, E.; Svedersky, L.P.; Finkle, B.S.; Palladino, M.A. Activation of human polymorphonuclear neutrophil functions by interferon-gamma and tumor necrosis factors. J. Immunol. 1985, 135, 2069–2073. [Google Scholar] [PubMed]
- Kucka, K.; Lang, I.; Zhang, T.; Siegmund, D.; Medler, J.; Wajant, H. Membrane lymphotoxin-α2β is a novel tumor necrosis factor (TNF) receptor 2 (TNFR2) agonist. Cell Death Dis. 2021, 12, 360. [Google Scholar] [CrossRef] [PubMed]
- Gubernatorova, E.O.; Polinova, A.I.; Petropavlovskiy, M.M.; Namakanova, O.A.; Medvedovskaya, A.D.; Zvartsev, R.V.; Telegin, G.B.; Drutskaya, M.S.; Nedospasov, S.A. Dual role of tnf and ltα in carcinogenesis as implicated by studies in mice. Cancers 2021, 13, 1775. [Google Scholar] [CrossRef]
- Roach, D.R.; Briscoe, H.; Saunders, B.; France, M.P.; Riminton, S.; Britton, W.J. Secreted lymphotoxin-alpha is essential for the control of an intracellular bacterial infection. J. Exp. Med. 2001, 193, 239–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruglov, A.A.; Grivennikov, S.I.; Kuprash, D.V.; Winsauer, C.; Prepens, S.; Seleznik, G.M.; Eberl, G.; Littman, D.R.; Heikenwalder, M.; Tumanov, A.V.; et al. Nonredundant function of soluble ltα3 produced by innate lymphoid cells in intestinal homeostasis. Science 2013, 342, 1243–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Etemadi, N.; Holien, J.K.; Chau, D.; Dewson, G.; Murphy, J.M.; Alexander, W.S.; Parker, M.W.; Silke, J.; Nachbur, U. Lymphotoxin α induces apoptosis, necroptosis and inflammatory signals with the same potency as tumour necrosis factor. FEBS J. 2013, 280, 5283–5297. [Google Scholar] [CrossRef]
- Pegoretti, V.; Baron, W.; Laman, J.D.; Eisel, U.L.M. Selective Modulation of TNF–TNFRs Signaling: Insights for Multiple Sclerosis Treatment. Front. Immunol. 2018, 9, 925. [Google Scholar] [CrossRef]
- Wajant, H.; Siegmund, D. TNFR1 and TNFR2 in the Control of the Life and Death Balance of Macrophages. Front. Cell Dev. Biol. 2019, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Raeiszadeh, M.; Verney, M.; Craddock, C.; Wajant, H.; Moss, P.; Chen, F. TNFR2 Is Expressed Preferentially By Late Differentiated CD8 T-Cells and Can be Triggered By TNFR2-Specific Ligand to Induce Cell Death of Recently Activated Antigen-Specific T Cells: A Possible Role of TNFR2 in T-Cell Deflation. Blood 2014, 124, 4352. [Google Scholar] [CrossRef]
- Hijdra, D.; Vorselaars, A.D.; Grutters, J.C.; Claessen, A.M.; Rijkers, G.T. Differential expression of TNFR1 (CD120a) and TNFR2 (CD120b) on subpopulations of human monocytes. J. Inflamm. 2012, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Beldi, G.; Bahiraii, S.; Lezin, C.; Nouri Barkestani, M.; Abdelgawad, M.E.; Uzan, G.; Naserian, S. TNFR2 Is a Crucial Hub Controlling Mesenchymal Stem Cell Biological and Functional Properties. Front. Cell Dev. Biol. 2020, 8, 596831. [Google Scholar] [CrossRef] [PubMed]
- Naserian, S.; Shamdani, S.; Arouche, N.; Uzan, G. Regulatory T cell induction by mesenchymal stem cells depends on the expression of TNFR2 by T cells. Stem Cell Res. Ther. 2020, 11, 534. [Google Scholar] [CrossRef] [PubMed]
- Morton, P.E.; Perrin, C.; Levitt, J.; Matthews, D.R.; Marsh, R.J.; Pike, R.; McMillan, D.; Maloney, A.; Poland, S.; Ameer-Beg, S.; et al. TNFR1 membrane reorganization promotes distinct modes of TNFα signaling. Sci. Signal. 2019, 12, 2418. [Google Scholar] [CrossRef] [PubMed]
- Cabal-Hierro, L.; Lazo, P.S. Signal transduction by tumor necrosis factor receptors. Cell. Signal. 2012, 24, 1297–1305. [Google Scholar] [CrossRef] [PubMed]
- Micheau, O.; Tschopp, J. Induction of TNF Receptor I-Mediated Apoptosis via Two Sequential Signaling Complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Zhao, H.; Jin, M.; Zhu, H.; Shan, B.; Geng, J.; Dziedzic, S.A.; Amin, P.; Mifflin, L.; Naito, M.G.; et al. Modulating TRADD to restore cellular homeostasis and inhibit apoptosis. Nature 2020, 587, 133–138. [Google Scholar] [CrossRef]
- Fischer, R.; Kontermann, R.E.; Pfizenmaier, K. Selective Targeting of TNF Receptors as a Novel Therapeutic Approach. Front. Cell Dev. Biol. 2020, 8, 401. [Google Scholar] [CrossRef]
- Van Ostade, X.; Vandenabeele, P.; Everaerdt, B.; Loetscher, H.; Gentz, R.; Brockhaus, M.; Lesslauer, W.; Tavernier, J.; Brouckaert, P.; Fiers, W. Human TNF mutants with selective activity on the p55 receptor. Nature 1993, 361, 266–269. [Google Scholar] [CrossRef]
- Barbara, J.A.; Smith, W.B.; Gamble, J.R.; Van Ostade, X.; Vandenabeele, P.; Tavernier, J.; Fiers, W.; Vadas, M.A.; Lopez, A.F. Dissociation of TNF-alpha cytotoxic and proinflammatory activities by p55 receptor- and p75 receptor-selective TNF-alpha mutants. EMBO J. 1994, 13, 843–850. [Google Scholar] [CrossRef]
- Loetscher, H.; Stueber, D.; Banner, D.; Mackay, F.; Lesslauer, W. Human tumor necrosis factor alpha (TNF alpha) mutants with exclusive specificity for the 55-kDa or 75-kDa TNF receptors. J. Biol. Chem. 1993, 268, 26350–26357. [Google Scholar] [CrossRef]
- Hube, F.; Hauner, H. The two tumor necrosis factor receptors mediate opposite effects on differentiation and glucose metabolism in human adipocytes in primary culture. Endocrinology 2000, 141, 2582–2588. [Google Scholar] [CrossRef]
- Lees, D.M.; Pallikaros, Z.; Corder, R. The p55 tumor necrosis factor receptor (CD120a) induces endothelin-1 synthesis in endothelial and epithelial cells. Eur. J. Pharmacol. 2000, 390, 89–94. [Google Scholar] [CrossRef]
- Shikama, H.; Miyata, K.; Sakae, N.; Mitsuishi, Y.; Nishimura, K.; Kuroda, K.; Kato, M. Novel mutein of tumor necrosis factor alpha (F4614) with reduced hypotensive effect. J. Interferon Cytokine Res. 1995, 15, 677–684. [Google Scholar] [CrossRef]
- Atarashi, Y.; Yasumura, S.; Nambu, S.; Yoshio, Y.; Murakami, J.; Takahara, T.; Higuchi, K.; Watanabe, A.; Miyata, K.; Kato, M. A novel human tumor necrosis factor alfa mutein, F4614, inhibits in vitro and in vivo growth of murine and human hepatoma: Implication for immunotherapy of human hepatocellular carcinoma. Hepatology 1998, 28, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Cha, S.-S.; Kim, J.-S.; Cho, H.-S.; Shin, N.-K.; Jeong, W.; Shin, H.-C.; Kim, Y.J.; Hahn, J.H.; Oh, B.-H. High Resolution Crystal Structure of a Human Tumor Necrosis Factor-α Mutant with Low Systemic Toxicity. J. Biol. Chem. 1998, 273, 2153–2160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munoz Pinto, M.F.; Campbell, S.J.; Simoglou Karali, C.; Johanssen, V.A.; Bristow, C.; Cheng, V.W.T.; Zarghami, N.; Larkin, J.R.; Pannell, M.; Hearn, A.; et al. Selective blood-brain barrier permeabilization of brain metastases by a type 1 receptor-selective tumor necrosis factor mutein. Neuro-Oncology 2021, 24, 52–63. [Google Scholar] [CrossRef]
- Van Ostade, X.; Tavernier, J.; Prangé, T.; Fiers, W. Localization of the active site of human tumour necrosis factor (hTNF) by mutational analysis. EMBO J. 1991, 10, 827–836. [Google Scholar] [CrossRef] [PubMed]
- Mayes, P.A.; Hance, K.W.; Hoos, A. The promise and challenges of immune agonist antibody development in cancer. Nat. Rev. Drug Discov. 2018, 17, 509–527. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.U.; Shin, W.; Son, J.Y.; Yoo, K.-Y.; Heo, Y.-S. Molecular Basis for the Neutralization of Tumor Necrosis Factor α by Certolizumab Pegol in the Treatment of Inflammatory Autoimmune Diseases. Int. J. Mol. Sci. 2017, 18, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genovese, M.C.; Cohen, S.; Moreland, L.; Lium, D.; Robbins, S.; Newmark, R.; Bekker, P. Combination Therapy with Etanercept and Anakinra in the Treatment of Patients with Rheumatoid Arthritis Who Have Been Treated Unsuccessfully with Methotrexate. Arthritis Rheum. 2004, 50, 1412–1419. [Google Scholar] [CrossRef]
- Goodall, L.J.; Ovecka, M.; Rycroft, D.; Friel, S.L.; Sanderson, A.; Mistry, P.; Davies, M.L.; Stoop, A.A. Pharmacokinetic and pharmacodynamic characterisation of an anti-mouse TNF receptor 1 domain antibody formatted for in vivo half-life extension. PLoS ONE 2015, 10, e0137065. [Google Scholar] [CrossRef] [Green Version]
- Gouweleeuw, L.; Wajant, H.; Maier, O.; Eisel, U.L.M.; Blankesteijn, W.M.; Schoemaker, R.G. Effects of selective TNFR1 inhibition or TNFR2 stimulation, compared to non-selective TNF inhibition, on (neuro)inflammation and behavior after myocardial infarction in male mice. Brain Behav. Immun. 2021, 93, 156–171. [Google Scholar] [CrossRef] [PubMed]
- McCann, F.E.; Perocheau, D.P.; Ruspi, G.; Blazek, K.; Davies, M.L.; Feldmann, M.; Dean, J.L.E.; Stoop, A.A.; Williams, R.O. Selective tumor necrosis factor receptor i blockade is antiinflammatory and reveals immunoregulatory role of tumor necrosis factor receptor II in collagen-induced arthritis. Arthritis Rheumatol. 2014, 66, 2728–2738. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.K.; Maier, O.; Fischer, R.; Fairless, R.; Hochmeister, S.; Stojic, A.; Pick, L.; Haar, D.; Musiol, S.; Storch, M.K.; et al. Antibody-mediated inhibition of TNFR1 attenuates disease in a mouse model of multiple sclerosis. PLoS ONE 2014, 9, 90117. [Google Scholar] [CrossRef] [PubMed]
- Zettlitz, K.A.; Lorenz, V.; Landauer, K.; Münkel, S.; Herrmann, A.; Scheurich, P.; Pfizenmaier, K.; Kontermann, R. ATROSAB, a humanized antagonistic anti-tumor necrosis factor receptor one-specific antibody. MAbs 2010, 2, 639–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, F.; Liebig, T.; Guenzi, E.; Herrmann, A.; Scheurich, P.; Pfizenmaier, K.; Kontermann, R.E. Antagonistic TNF Receptor One-Specific Antibody (ATROSAB): Receptor Binding and In Vitro Bioactivity. PLoS ONE 2013, 8, e72156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Feng, Z.; Wang, Y.; Ma, S.; Hu, Z.; Yang, P.; Chai, Y.; Xie, X. Discovery of Novel Ligands for TNF-α and TNF Receptor-1 through Structure-Based Virtual Screening and Biological Assay. J. Chem. Inf. Model. 2017, 57, 1101–1111. [Google Scholar] [CrossRef]
- Saddala, M.S.; Huang, H. Identification of novel inhibitors for TNFα, TNFR1 and TNFα-TNFR1 complex using pharmacophore-based approaches. J. Transl. Med. 2019, 17, 215. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.H.; Schaaf, T.M.; Grant, B.D.; Lim, C.K.W.; Bawaskar, P.; Aldrich, C.C.; Thomas, D.D.; Sachs, J.N. Noncompetitive inhibitors of TNFR1 probe conformational activation states. Sci. Signal. 2019, 12, 5637. [Google Scholar] [CrossRef]
- Van De Kar, N.C.A.J.; Kooistra, T.; Vermeer, M.; Lesslauer, W.; Monnens, L.A.H.; Van Hinsbergh, V.W.M. Tumor necrosis factor α induces endothelial galactosyl transferase activity and verocytotoxin receptors. Role of specific tumor necrosis factor receptors and protein kinase C. Blood 1995, 85, 734–743. [Google Scholar] [CrossRef]
- Yui, J.; Hemmings, D.; Garcia-Lloret, M.; Guilbert, L.J. Expression of the human p55 and p75 tumor necrosis factor receptors in primary villous trophoblasts and their role in cytotoxic signal transduction. Biol. Reprod. 1996, 55, 400–409. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Garcia-Lloret, M.; Winkler-Lowen, B.; Miller, R.; Simpson, K.; Guilbert, L.J. ICAM-1-mediated adhesion of peripheral blood monocytes to the maternal surface of placental syncytiotrophoblasts: Implications for placental villitis. Am. J. Pathol. 1997, 150, 1845–1860. [Google Scholar] [PubMed]
- Dong, Y.; Fischer, R.; Naudé, P.J.W.; Maier, O.; Nyakas, C.; Duffey, M.; Van Der Zee, E.A.; Dekens, D.; Douwenga, W.; Herrmann, A.; et al. Essential protective role of tumor necrosis factor receptor 2 in neurodegeneration. Proc. Natl. Acad. Sci. USA 2016, 113, 12304–12309. [Google Scholar] [CrossRef] [Green Version]
- Fischer, R.; Proske, M.; Duffey, M.; Stangl, H.; Martinez, G.F.; Peters, N.; Kraske, A.; Straub, R.H.; Bethea, J.R.; Kontermann, R.E.; et al. Selective Activation of Tumor Necrosis Factor Receptor II Induces Antiinflammatory Responses and Alleviates Experimental Arthritis. Arthritis Rheumatol. 2018, 70, 722–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, M.; Biehl, M.; Steinfatt, T.; Brandl, A.; Kums, J.; Amich, J.; Vaeth, M.; Kuen, J.; Holtappels, R.; Podlech, J.; et al. Exogenous TNFR2 activation protects from acute GvHD via host T reg cell expansion. J. Exp. Med. 2016, 213, 1881–1900. [Google Scholar] [CrossRef] [PubMed]
- Fischer, R.; Maier, O.; Siegemund, M.; Wajant, H.; Scheurich, P.; Pfizenmaier, K. A TNF receptor 2 selective agonist rescues human neurons from oxidative stress-induced cell death. PLoS ONE 2011, 6, e27621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiley, S.R.; Schooley, K.; Smolak, P.J.; Din, W.S.; Huang, C.P.; Nicholl, J.K.; Sutherland, G.R.; Smith, T.D.; Rauch, C.; Smith, C.A.; et al. Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity 1995, 3, 673–682. [Google Scholar] [CrossRef] [Green Version]
- Diaz Arguello, O.A.; Haisma, H.J.; Apoptosis-Inducing, H. Apoptosis-Inducing TNF Superfamily Ligands for Cancer Therapy. Cancers 2021, 13, 1543. [Google Scholar] [CrossRef]
- Zhang, B.; van Roosmalen, I.A.M.; Reis, C.R.; Setroikromo, R.; Quax, W.J. Death receptor 5 is activated by fucosylation in colon cancer cells. FEBS J. 2019, 286, 555–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, W.; Immunology, P.H. TNF-related apoptosis-inducing ligand (TRAIL) induces apoptosis in Fas ligand-resistant melanoma cells and mediates CD4 T cell killing of target cells. Am. Assoc. Immnol. 1998, 161, 2195–2200. [Google Scholar]
- Mori, S.; Murakami-Mori, K.; Nakamura, S.; Ashkenazi, A.; Bonavida, B. Sensitization of AIDS-Kaposi’s sarcoma cells to Apo-2 ligand-induced apoptosis by actinomycin D. J. Immunol. 1999, 162, 5616–56123. [Google Scholar] [PubMed]
- Rieger, J.; Naumann, U.; Glaser, T.; Ashkenazi, A.; Weller, M. APO2 ligand: A novel lethal weapon against malignant glioma? FEBS Lett. 1998, 427, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Walczak, H.; Miller, R.E.; Ariail, K.; Gliniak, B.; Griffith, T.S.; Kubin, M.; Chin, W.; Jones, J.; Woodward, A.; Le, T.; et al. Tumoricidal activity of tumor necrosis factor–related apoptosis–inducing ligand in vivo. Nat. Med. 1999, 5, 157–163. [Google Scholar] [CrossRef]
- Ashkenazi, A.; Pai, R.C.; Fong, S.; Leung, S.; Lawrence, D.A.; Marsters, S.A.; Blackie, C.; Chang, L.; McMurtrey, A.E.; Hebert, A.; et al. Dulanermin Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Investig. 1999, 104, 155–162. [Google Scholar] [CrossRef] [PubMed]
- Zuch de Zafra, C.L.; Ashkenazi, A.; Darbonne, W.C.; Cheu, M.; Totpal, K.; Ortega, S.; Flores, H.; Walker, M.D.; Kabakoff, B.; Lum, B.L.; et al. Antitherapeutic antibody-mediated hepatotoxicity of recombinant human Apo2L/TRAIL in the cynomolgus monkey. Cell Death Dis. 2016, 7, e2338. [Google Scholar] [CrossRef] [Green Version]
- Kelley, S.K.; Harris, L.A.; Xie, D.; Deforge, L.; Totpal, K.; Bussiere, J.; Fox, J.A. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: Characterization of in vivo efficacy, pharmacokinetics, and safety. J. Pharmacol. Exp. Ther. 2001, 299, 31–38. [Google Scholar] [PubMed]
- Snajdauf, M.; Havlova, K.; Vachtenheim, J.; Ozaniak, A.; Lischke, R.; Bartunkova, J.; Smrz, D.; Strizova, Z. The TRAIL in the Treatment of Human Cancer: An Update on Clinical Trials. Front. Mol. Biosci. 2021, 8, 87. [Google Scholar] [CrossRef] [PubMed]
- Reis, C.R.; van der Sloot, A.M.; Szegezdi, E.; Natoni, A.; Tur, V.; Cool, R.H.; Samali, A.; Serrano, L.; Quax, W.J. Enhancement of Antitumor Properties of rhTRAIL by Affinity Increase toward Its Death Receptors. Biochemistry 2009, 48, 2180–2191. [Google Scholar] [CrossRef]
- Huang, M.; Zhu, H.; Yi, C.; Yan, J.; Wei, L.; Yang, X.; Chen, S.; Huang, Y. A novel TRAIL mutant-TRAIL-Mu3 enhances the antitumor effects by the increased affinity and the up-expression of DR5 in pancreatic cancer. Cancer Chemother. Pharmacol. 2018, 82, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Kelley, R.F.; Totpal, K.; Lindstrom, S.H.; Mathieu, M.; Billeci, K.; DeForge, L.; Pai, R.; Hymowitz, S.G.; Ashkenazi, A. Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of Death Receptor (DR) 5 than DR4 to apoptosis signaling. J. Biol. Chem. 2005, 280, 2205–2212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacFarlane, M.; Kohlhaas, S.L.; Sutcliffe, M.J.; Dyer, M.J.S.; Cohen, G.M. TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res. 2005, 65, 11265–11270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tur, V.; van der Sloot, A.M.; Reis, C.R.; Szegezdi, E.; Cool, R.H.; Samali, A.; Serrano, L.; Quax, W.J. DR4-selective tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) variants obtained by structure-based design. J. Biol. Chem. 2008, 283, 20560–20568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, R.; Albarenque, S.M.; Cool, R.H.; Quax, W.J.; Mohr, A.; Zwacka, R.M. DR4 specific TRAIL variants are more efficacious than wild-type TRAIL in pancreatic cancer. Cancer Biol. Ther. 2014, 15, 1658–1666. [Google Scholar] [CrossRef] [Green Version]
- Reis, C.R.; Van Der Sloot, A.M.; Natoni, A.; Szegezdi, E.; Setroikromo, R.; Meijer, M.; Sjollema, K.; Stricher, F.; Cool, R.H.; Samali, A.; et al. Rapid and efficient cancer cell killing mediated by high-affinity death receptor homotrimerizing TRAIL variants. Cell Death Dis. 2010, 1, e83. [Google Scholar] [CrossRef] [Green Version]
- Szegezdi, E.; Reis, C.R.; Der Sloot, A.M.V.; Natoni, A.; O’Reilly, A.; Reeve, J.; Cool, R.H.; O’Dwyer, M.; Knapper, S.; Serrano, L.; et al. Targeting AML through DR4 with a novel variant of rhTRAIL. J. Cell. Mol. Med. 2011, 15, 2216–2231. [Google Scholar] [CrossRef] [Green Version]
- Reis, C.R.; van Assen, A.H.G.; Quax, W.J.; Cool, R.H. Unraveling the Binding Mechanism of Trivalent Tumor Necrosis Factor Ligands and Their Receptors. Mol. Cell. Proteom. 2011, 10, M110.002808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gasparian, M.E.; Chernyak, B.V.; Dolgikh, D.A.; Yagolovich, A.V.; Popova, E.N.; Sycheva, A.M.; Moshkovskii, S.A.; Kirpichnikov, M.P. Generation of new TRAIL mutants DR5-A and DR5-B with improved selectivity to death receptor 5. Apoptosis 2009, 14, 778–787. [Google Scholar] [CrossRef]
- Van Der Sloot, A.M.; Tur, V.; Szegezdi, E.; Mullally, M.M.; Cool, R.H.; Samali, A.; Serrano, L.; Quax, W.J. Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc. Natl. Acad. Sci. USA 2006, 103, 8634–8639. [Google Scholar] [CrossRef] [Green Version]
- van Roosmalen, I.A.M.; Reis, C.R.; Setroikromo, R.; Yuvaraj, S.; Joseph, J.V.; Tepper, P.G.; Kruyt, F.A.E.; Quax, W.J. The ER stress inducer DMC enhances TRAIL-induced apoptosis in glioblastoma. Springerplus 2014, 3, 495. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Zijlstra, S.N.; Soto-Gamez, A.; Setroikromo, R.; Quax, W.J. Artemisinin Derivatives Stimulate DR5-Specific TRAIL-Induced Apoptosis by Regulating Wildtype P53. Cancers 2020, 12, 2514. [Google Scholar] [CrossRef]
- Zhou, X.; Soto-Gamez, A.; Nijdam, F.; Setroikromo, R.; Quax, W.J. Dihydroartemisinin-transferrin adducts enhance TRAIL-induced apoptosis in triple-negative breast cancer in a P53-independent and ROS-dependent manner. Front. Oncol. 2022, 11, 5452. [Google Scholar] [CrossRef]
- Soto-Gamez, A.; Wang, Y.; Zhou, X.; Seras, L.; Quax, W.; Demaria, M. Enhanced extrinsic apoptosis of therapy-induced senescent cancer cells using a death receptor 5 (DR5) selective agonist. Cancer Lett. 2022, 525, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Arabpour, M.; Cool, R.H.; Faber, K.N.; Quax, W.J.; Haisma, H.J. Receptor-specific TRAIL as a means to achieve targeted elimination of activated hepatic stellate cells. J. Drug Target. 2017, 25, 360–369. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, B.; Chen, D.; Setroikromo, R.; Haisma, H.J.; Quax, W.J. Histone Deacetylase Inhibitors Sensitize TRAIL-Induced Apoptosis in Colon Cancer Cells. Cancers 2019, 11, 645. [Google Scholar] [CrossRef] [Green Version]
- Nelson, C.A.; Warren, J.T.; Wang, M.W.H.; Teitelbaum, S.L.; Fremont, D.H. RANKL employs distinct binding modes to engage RANK and the osteoprotegerin decoy receptor. Structure 2012, 20, 1971–1982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, J.; Cronin, S.J.F.; Penninger, J.M. Targeting the RANKL/RANK/OPG Axis for Cancer Therapy. Front. Oncol. 2020, 10, 1283. [Google Scholar] [CrossRef] [PubMed]
- Galibert, L.; Tometsko, M.E.; Andersen, D.M.; Cosman, D.; Dougall, W.C. The involvement of multiple tumor necrosis factor receptor (TNFR)- associated factors in the signaling mechanisms of receptor activator of NF- κB, a member of the TNFR superfamily. J. Biol. Chem. 1998, 273, 34120–34127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ilchovska, D.; Barrow, D.M. An Overview of the NF-kB mechanism of pathophysiology in rheumatoid arthritis, investigation of the NF-kB ligand RANKL and related nutritional interventions. Autoimmun. Rev. 2021, 20, 102741. [Google Scholar] [CrossRef]
- Verma, D.; Zanetti, C.; Godavarthy, P.S.; Kumar, R.; Minciacchi, V.R.; Pfeiffer, J.; Metzler, M.; Lefort, S.; Maguer-Satta, V.; Nicolini, F.E.; et al. Bone marrow niche-derived extracellular matrix-degrading enzymes influence the progression of B-cell acute lymphoblastic leukemia. Leukemia 2020, 34, 1540–1552. [Google Scholar] [CrossRef] [PubMed]
- Herrera, J.; Henke, C.A.; Bitterman, P.B. Extracellular matrix as a driver of progressive fibrosis. J. Clin. Investig. 2018, 128, 45–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habibie, H.; Adhyatmika, A.; Schaafsma, D.; Melgert, B.N. The role of osteoprotegerin (OPG) in fibrosis: Its potential as a biomarker and/or biological target for the treatment of fibrotic diseases. Pharmacol. Ther. 2021, 228, 107941. [Google Scholar] [CrossRef] [PubMed]
- Hymowitz, S.G.; Christinger, H.W.; Fuh, G.; Ultsch, M.; O’Connell, M.; Kelley, R.F.; Ashkenazi, A.; De Vos, A.M. Triggering cell death: The crystal structure of Apo2L/TRAIL in a complex with death receptor 5. Mol. Cell 1999, 4, 563–571. [Google Scholar] [CrossRef]
- Banner, D.W.; D’Arcy, A.; Janes, W.; Gentz, R.; Schoenfeld, H.J.; Broger, C.; Loetscher, H.; Lesslauer, W. Crystal structure of the soluble human 55 kd TNF receptor-human TNFβ complex: Implications for TNF receptor activation. Cell 1993, 73, 431–445. [Google Scholar] [CrossRef]
- Lam, J.; Nelson, C.A.; Ross, F.P.; Teitelbaum, S.L.; Fremont, D.H. Crystal structure of the TRANCE/RANKL cytokine reveals determinants of receptor-ligand specificity. J. Clin. Investig. 2001, 108, 971–979. [Google Scholar] [CrossRef]
- Wang, Y.; van Assen, A.H.G.; Reis, C.R.; Setroikromo, R.; van Merkerk, R.; Boersma, Y.L.; Cool, R.H.; Quax, W.J. Novel RANKL DE-loop mutants antagonize RANK-mediated osteoclastogenesis. FEBS J. 2017, 284, 2501–2512. [Google Scholar] [CrossRef] [Green Version]
- Wassenaar, T.A.; Quax, W.J.; Mark, A.E. The conformation of the extracellular binding domain of Death Receptor 5 in the presence and absence of the activating ligand TRAIL: A molecular dynamics study. Proteins Struct. Funct. Bioinform. 2008, 70, 333–343. [Google Scholar] [CrossRef]
- Neumann, S.; Bidon, T.; Branschädel, M.; Krippner-Heidenreich, A.; Scheurich, P.; Doszczak, M. The transmembrane domains of TNF-related apoptosis-inducing ligand (TRAIL) receptors 1 and 2 co-regulate apoptotic signaling capacity. PLoS ONE 2012, 7, e42526. [Google Scholar] [CrossRef]
- Cheng, T.; Pavlos, N.J.; Wang, C.; Tan, J.W.Y.; Lin, J.M.; Cornish, J.; Zheng, M.H.; Xu, J. Mutations within the TNF-like core domain of RANKL impair osteoclast differentiation and activation. Mol. Endocrinol. 2009, 23, 35–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Michiels, T.; Setroikromo, R.; van Merkerk, R.; Cool, R.H.; Quax, W.J. Creation of RANKL mutants with low affinity for decoy receptor OPG and their potential anti-fibrosis activity. FEBS J. 2019, 286, 3582–3593. [Google Scholar] [CrossRef]
- Ambroszkiewicz, J.; Sands, D.; Gajewska, J.; Chelchowska, M.; Laskowska-Klita, T. Bone turnover markers, osteoprotegerin and RANKL cytokines in children with cystic fibrosis. Adv. Med. Sci. 2013, 58, 338–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Heron, L.; Guillaume, C.; Velard, F.; Braux, J.; Touqui, L.; Moriceau, S.; Sermet-Gaudelus, I.; Laurent-Maquin, D.; Jacquot, J. Cystic fibrosis transmembrane conductance regulator (CFTR) regulates the production of osteoprotegerin (OPG) and prostaglandin (PG) E2 in human bone. J. Cyst. Fibros. 2010, 9, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Toffoli, B.; Pickering, R.J.; Tsorotes, D.; Wang, B.; Bernardi, S.; Kantharidis, P.; Fabris, B.; Zauli, G.; Secchiero, P.; Thomas, M.C. Osteoprotegerin promotes vascular fibrosis via a TGF-β1 autocrine loop. Atherosclerosis 2011, 218, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Adhyatmika, A.; Putri, K.S.S.S.; Beljaars, L.; Melgert, B.N. The elusive antifibrotic macrophage. Front. Med. 2015, 2, 81. [Google Scholar] [CrossRef] [Green Version]
- Kurinami, H.; Shimamura, M.; Nakagami, H.; Shimizu, H.; Koriyama, H.; Kawano, T.; Wakayama, K.; Mochizuki, H.; Rakugi, H.; Morishita, R. A Novel Therapeutic Peptide as a Partial Agonist of RANKL in Ischemic Stroke. Sci. Rep. 2016, 6, 38062. [Google Scholar] [CrossRef] [Green Version]
- Aoki, K.; Saito, H.; Itzstein, C.; Ishiguro, M.; Shibata, T.; Blanque, R.; Mian, A.H.; Takahashi, M.; Suzuki, Y.; Yoshimatsu, M.; et al. A TNF receptor loop peptide mimic blocks RANK ligand–induced signaling, bone resorption, and bone loss. J. Clin. Investig. 2006, 116, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Ta, H.M.; Nguyen, G.T.T.; Jin, H.M.; Choi, J.; Park, H.; Kim, N.; Hwang, H.Y.; Kim, K.K. Structure-based development of a receptor activator of nuclear factor-κB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis. Proc. Natl. Acad. Sci. USA 2010, 107, 20281–20286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quax, W.J.; Van Assen, A.H.G.; Wang, Y.Z. TNF-family member Receptor Activator of NF-κB (RANK) and RANK-Ligand (RANKL) in bone remodelling. In Proceedings of the IOP Conference Series: Earth and Environmental Science, Hangzhou, China, 26–29 June 2018; IOP Publishing: Bristol, UK, 2018; Volume 185, p. 012001. [Google Scholar]
- Denosumab in Treating Patients With Bone Loss Due to Donor Stem Cell Transplant. Available online: https://clinicaltrials.gov/ct2/show/NCT03925532 (accessed on 5 January 2022).
- The Effects of 12-Months of Denosumab on Bone Density in Prevalent Kidney Transplant Recipients. Available online: https://clinicaltrials.gov/ct2/show/NCT03960554 (accessed on 5 January 2022).
- Efficacy of Denosumab on Normal BMD in Women Receiving Adjuvant Aromatase Inhibitors for Early Breast Cancer. Available online: https://www.clinicaltrials.gov/ct2/show/NCT03324932 (accessed on 5 January 2022).
- Templeton, A.J.; Stalder, L.; Bernhard, J.; Brauchli, P.; Gillessen, S.; Hayoz, S.; Klingbiel, D.; Matter-Walstra, K.; Thurlimann, B.J.K.; Von Moos, R. Prevention of symptomatic skeletal events with denosumab administered every 4 weeks versus every 12 weeks: A noninferiority phase III trial (SAKK 96/12, REDUSE). J. Clin. Oncol. 2014, 32, TPS5095. [Google Scholar] [CrossRef]
- Yamada, A.; Arakaki, R.; Saito, M.; Kudo, Y.; Ishimaru, N. Dual role of Fas/FasL-mediated signal in peripheral immune tolerance. Front. Immunol. 2017, 8, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Q.; Fu, T.M.; Cruz, A.C.; Sengupta, P.; Thomas, S.K.; Wang, S.; Siegel, R.M.; Wu, H.; Chou, J.J. Structural Basis and Functional Role of Intramembrane Trimerization of the Fas/CD95 Death Receptor. Mol. Cell 2016, 61, 602–613. [Google Scholar] [CrossRef] [Green Version]
- Huang, B.; Eberstadt, M.; Olejniczak, E.T.; Meadows, R.P.; Feslk, S.W. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature 1996, 384, 638–641. [Google Scholar] [CrossRef]
- Liu, W.; Ramagopal, U.; Cheng, H.; Bonanno, J.B.; Toro, R.; Bhosle, R.; Zhan, C.; Almo, S.C. Crystal Structure of the Complex of Human FasL and Its Decoy Receptor DcR3. Structure 2016, 24, 2016–2023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Zhao, Y.; Wang, C.; Ji, H.; Yu, J.; Liu, C.; Liu, A. A novel synthetic chitosan selenate (CS) induces apoptosis in A549 lung cancer cells via the Fas/FasL pathway. Int. J. Biol. Macromol. 2020, 158, 689–697. [Google Scholar] [CrossRef] [PubMed]
- Bellesi, S.; Metafuni, E.; Hohaus, S.; Maiolo, E.; Marchionni, F.; D’Innocenzo, S.; La Sorda, M.; Ferraironi, M.; Ramundo, F.; Fantoni, M.; et al. Increased CD95 (Fas) and PD-1 expression in peripheral blood T lymphocytes in COVID-19 patients. Br. J. Haematol. 2020, 191, 207–211. [Google Scholar] [CrossRef]
- Mitsiades, N.; Poulaki, V.; Mitsiades, C.S.; Koutras, D.A.; Chrousos, G.P. Apoptosis induced by FasL and TRAIL/Apo2L in the pathogenesis of thyroid diseases. Trends Endocrinol. Metab. 2001, 12, 384–390. [Google Scholar] [CrossRef]
- Jimbo, H.; Nagai, H.; Fujiwara, S.; Shimoura, N.; Nishigori, C. Fas-FasL interaction in cytotoxic T cell-mediated vitiligo: The role of lesional expression of tumor necrosis factor-α and interferon-γ in Fas-mediated melanocyte apoptosis. Exp. Dermatol. 2020, 29, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Kouzmenko, A.; Kato, S. Targeting Fas/FasL signaling, a new strategy for maintaining bone health. Expert Opin. Ther. Targets 2011, 15, 1143–1145. [Google Scholar] [CrossRef] [PubMed]
- Malleter, M.; Tauzin, S.; Bessede, A.; Castellano, R.; Goubard, A.; Godey, F.; Levêque, J.; Jézéquel, P.; Campion, L.; Campone, M.; et al. CD95L cell surface cleavage triggers a prometastatic signaling pathway in triple-negative breast cancer. Cancer Res. 2013, 73, 6711–6721. [Google Scholar] [CrossRef] [Green Version]
- Greaney, P.; Nahimana, A.; Lagopoulos, L.; Etter, A.L.; Aubry, D.; Attinger, A.; Beltraminelli, N.; Huni, B.; Bassi, I.; Sordat, B.; et al. A Fas agonist induces high levels of apoptosis in haematological malignancies. Leuk. Res. 2006, 30, 415–426. [Google Scholar] [CrossRef]
- Tsao, T.S.; Lodish, H.F.; Fruebis, J. ACRP30, a new hormone controlling fat and glucose metabolism. Eur. J. Pharmacol. 2002, 440, 213–221. [Google Scholar] [CrossRef]
- Eisele, G.; Roth, P.; Hasenbach, K.; Aulwurm, S.; Wolpert, F.; Tabatabai, G.; Wick, W.; Weller, M. APO010, a synthetic hexameric CD95 ligand, induces human glioma cell death in vitro and in vivo. Neuro-Oncology 2011, 13, 155–164. [Google Scholar] [CrossRef]
- Ocio, E.M.; Maiso, P.; Garayoa, M.; Dupuis, M.; Pandiella, A.; Miguel, J.F.S. The Activation of Fas Receptor by APO010, a Recombinant Form of Fas Ligand, Induces In Vitro and In Vivo Antimyeloma Activity. Blood 2007, 110, 1515. [Google Scholar] [CrossRef]
- Verbrugge, I.; Wissink, E.H.J.; Rooswinkel, R.W.; Jongsma, J.; Beltraminelli, N.; Dupuis, M.; Borst, J.; Verheij, M. Combining radiotherapy with APO010 in cancer treatment. Clin. Cancer Res. 2009, 15, 2031–2038. [Google Scholar] [CrossRef] [Green Version]
- A Phase I Dose Finding Study of APO010 in Patients With Solid Tumors. Available online: http://clinicaltrials.gov/show/NCT00437736 (accessed on 5 January 2022).
- Huang, J.H.; Tykocinski, M.L. CTLA-4-Fas ligand functions as a trans signal converter protein in bridging antigen-presenting cells and T cells. Int. Immunol. 2001, 13, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Georgopoulos, N.T.; Steele, L.P.; Thomson, M.J.; Selby, P.J.; Southgate, J.; Trejdosiewicz, L.K. A novel mechanism of CD40-induced apoptosis of carcinoma cells involving TRAF3 and JNK/AP-1 activation. Cell Death Differ. 2006, 13, 1789–1801. [Google Scholar] [CrossRef]
- Dranitzki-Elhalel, M.; Huang, J.H.; Sasson, M.; Rachmilewitz, J.; Parnas, M.; Tykocinski, M.L. CD40·FasL inhibits human T cells: Evidence for an auto-inhibitory loop-back mechanism. Int. Immunol. 2007, 19, 355–363. [Google Scholar] [CrossRef]
- Orbach, A.; Rachmilewitz, J.; Shani, N.; Isenberg, Y.; Parnas, M.; Huang, J.H.; Tykocinski, M.L.; Dranitzki-Elhalel, M. CD40·FasL and CTLA-4·FasL fusion proteins induce apoptosis in malignant cell lines by dual signaling. Am. J. Pathol. 2010, 177, 3159–3168. [Google Scholar] [CrossRef]
- Ahamadi-Fesharaki, R.; Fateh, A.; Vaziri, F.; Solgi, G.; Siadat, S.D.; Mahboudi, F.; Rahimi-Jamnani, F. Single-Chain Variable Fragment-Based Bispecific Antibodies: Hitting Two Targets with One Sophisticated Arrow. Mol. Ther.-Oncolytics 2019, 14, 38–56. [Google Scholar] [CrossRef] [Green Version]
- Samel, D.; Müller, D.; Gerspach, J.; Assohou-Luty, C.; Sass, G.; Tiegs, G.; Pfizenmaier, K.; Wajant, H. Generation of a FasL-based proapoptotic fusion protein devoid of systemic toxicity due to cell-surface antigen-restricted activation. J. Biol. Chem. 2003, 278, 32077–32082. [Google Scholar] [CrossRef] [Green Version]
- Villa-Morales, M.; Fernández-Piqueras, J. Targeting the Fas/FasL signaling pathway in cancer therapy. Expert Opin. Ther. Targets 2012, 16, 85–101. [Google Scholar] [CrossRef]
- Bremer, E.; Ten Cate, B.; Samplonius, D.F.; Mueller, N.; Wajant, H.; Stel, A.J.; Chamuleau, M.; Van De Loosdrecht, A.A.; Stieglmaier, J.; Fey, G.H.; et al. Superior activity of fusion protein scFvRit:sFasL over cotreatment with rituximab and fas agonists. Cancer Res. 2008, 68, 597–604. [Google Scholar] [CrossRef] [Green Version]
- Bremer, E.; Ten Cate, B.; Samplonius, D.F.; De Leij, L.F.M.H.; Helfrich, W. CD7-restricted activation of Fas-mediated apoptosis: A novel therapeutic approach for acute T-cell leukemia. Blood 2006, 107, 2863–2870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, D.V.; Sharma, R.; Ju, C.-Y.A.; Roffler, S.R.; Ju, S.-T. A recombinant scFv-FasLext as a targeting cytotoxic agent against human Jurkat-Ras cancer. J. Biomed. Sci. 2013, 20, 16. [Google Scholar] [CrossRef] [Green Version]
- Mauri, D.N.; Ebner, R.; Montgomery, R.I.; Kochel, K.D.; Cheung, T.C.; Yu, G.L.; Ruben, S.; Murphy, M.; Eisenberg, R.J.; Cohen, G.H.; et al. LIGHT, a new member of the TNF superfamily, and lymphotoxin α are ligands for herpesvirus entry mediator. Immunity 1998, 8, 21–30. [Google Scholar] [CrossRef] [Green Version]
- Brunetti, G.; Belisario, D.C.; Bortolotti, S.; Storlino, G.; Colaianni, G.; Faienza, M.F.; Sanesi, L.; Alliod, V.; Buffoni, L.; Centini, E.; et al. LIGHT/TNFSF14 Promotes Osteolytic Bone Metastases in Non-small Cell Lung Cancer Patients. J. Bone Miner. Res. 2020, 35, 671–680. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Zhan, C.; Cheng, H.; Kumar, P.R.; Bonanno, J.B.; Nathenson, S.G.; Almo, S.C. Mechanistic Basis for Functional Promiscuity in the TNF and TNF Receptor Superfamilies: Structure of the LIGHT:DcR3 Assembly. Structure 2014, 22, 1252–1262. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, M.T.; Dejardin, E.; dos Santos, N.R. Context-dependent roles for lymphotoxin-β receptor signaling in cancer development. Biochim. Biophys. Acta 2016, 1865, 204–219. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, R.; Garrett-Thomson, S.C.; Liu, W.; Almo, S.C.; Fiser, A. Redesigning HVEM Interface for Selective Binding to LIGHT, BTLA, and CD160. Structure 2020, 28, 1197–1205.e2. [Google Scholar] [CrossRef] [PubMed]
- Skeate, J.G.; Otsmaa, M.E.; Prins, R.; Fernandez, D.J.; Da Silva, D.M.; Kast, W.M. TNFSF14: LIGHTing the Way for Effective Cancer Immunotherapy. Front. Immunol. 2020, 11, 922. [Google Scholar] [CrossRef]
- Harrop, J.A.; McDonnell, P.C.; Brigham-Burke, M.; Lyn, S.D.; Minton, J.; Tan, K.B.; Dede, K.; Spampanato, J.; Silverman, C.; Hensley, P.; et al. Herpesvirus entry mediator ligand (HVEM-L), a novel ligand for HVEM/TR2, stimulates proliferation of T cells and inhibits HT29 cell growth. J. Biol. Chem. 1998, 273, 27548–27556. [Google Scholar] [CrossRef] [Green Version]
- Steinberg, M.W.; Shui, J.W.; Ware, C.F.; Kronenberg, M. Regulating the mucosal immune system: The contrasting roles of LIGHT, HVEM, and their various partners. Semin. Immunopathol. 2009, 31, 207–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heo, S.-K.; Ju, S.-A.; Lee, S.-C.; Park, S.-M.; Choe, S.-Y.; Kwon, B.; Kwon, B.S.; Kim, B.-S. LIGHT enhances the bactericidal activity of human monocytes and neutrophils via HVEM. J. Leukoc. Biol. 2006, 79, 330–338. [Google Scholar] [CrossRef] [Green Version]
- Holmes, T.D.; Wilson, E.B.; Black, E.V.I.; Benest, A.V.; Vaz, C.; Tan, B.; Tanavde, V.M.; Cook, G.P. Licensed human natural killer cells aid dendritic cell maturation via TNFSF14/LIGHT. Proc. Natl. Acad. Sci. USA 2014, 111, E5688–E5696. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Yu, P.; Wang, Y.; Wang, Y.; Fu, M.L.; Liu, W.; Sun, Y.; Fu, Y.X. NK-cell activation by LIGHT triggers tumor-specific CD8 + T-cell immunity to reject established tumors. Blood 2006, 107, 1342–1351. [Google Scholar] [CrossRef]
- Schneider, K.; Potter, K.G.; Ware, C.F. Lymphotoxin and LIGHT signaling pathways and target genes. Immunol. Rev. 2004, 202, 49–66. [Google Scholar] [CrossRef]
- Chen, M.C.; Hwang, M.J.; Chou, Y.C.; Chen, W.H.; Cheng, G.; Nakano, H.; Luh, T.Y.; Mai, S.C.; Hsieh, S.L. The role of apoptosis signal-regulating kinase 1 in lymphotoxin-β receptor-mediated cell death. J. Biol. Chem. 2003, 278, 16073–16081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, H.; Liang, C.; Ren, S.; Yue, C.; Wu, J. Prognostic value of DcR3 in solid tumors: A meta-analysis. Clin. Chim. Acta 2018, 481, 126–131. [Google Scholar] [CrossRef]
- Morishige, T.; Yoshioka, Y.; Inakura, H.; Tanabe, A.; Yao, X.; Tsunoda, S.; Tsutsumi, Y.; Mukai, Y.; Okada, N.; Nakagawa, S. Creation of a LIGHT mutant with the capacity to evade the decoy receptor for cancer therapy. Biomaterials 2010, 31, 3357–3363. [Google Scholar] [CrossRef] [PubMed]
- Rooney, I.A.; Butrovich, K.D.; Glass, A.A.; Borboroglu, S.; Benedict, C.A.; Whitbeck, J.C.; Cohen, G.H.; Eisenberg, R.J.; Ware, C.F. The lymphotoxin-β receptor is necessary and sufficient for LIGHT- mediated apoptosis of tumor cells. J. Biol. Chem. 2000, 275, 14307–14315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppensteiner, H.; Makowska, Z.; Volz, T.; et al. Specific and Nonhepatotoxic Degradation of Nuclear Hepatitis B Virus cccDNA. Science 2014, 343, 1221. [Google Scholar] [CrossRef] [PubMed]
- Riedl, T.; Faure-Dupuy, S.; Rolland, M.; Schuehle, S.; Hizir, Z.; Calderazzo, S.; Zhuang, X.; Wettengel, J.; Lopez, M.A.; Barnault, R.; et al. Hypoxia-Inducible Factor 1 Alpha–Mediated RelB/APOBEC3B Down-regulation Allows Hepatitis B Virus Persistence. Hepatology 2021, 74, 1766–1781. [Google Scholar] [CrossRef] [PubMed]
- Faure-Dupuy, S.; Riedl, T.; Rolland, M.; Hizir, Z.; Reisinger, F.; Neuhaus, K.; Schuehle, S.; Remouchamps, C.; Gillet, N.; Schönung, M.; et al. Control of APOBEC3B induction and cccDNA decay by NF-κB and miR-138-5p. JHEP Rep. 2021, 3, 100354. [Google Scholar] [CrossRef] [PubMed]




| Ligand | Specificity | Variants | Mutation Sites | Binding Affinity (nM) | Ref. | |
|---|---|---|---|---|---|---|
| TNFR1 | TNFR2 | |||||
| TNF-α | TNFR1 &TNFR2 | WT | / | 15.8 | 35.3 | [41] |
| TNFR1 | L29S | L29S | − 1 | − | [42] | |
| R32W | R32W | − | − | [42] | ||
| R32W-S86T | R32W/S86T | 3540 | NB 2 | [42] | ||
| F4614 | T5G/P6D/R29V | − | − | [38] | ||
| M3S | L29S/S52I/Y56F and 449/455del | − | − | [40] | ||
| mutTNF G4 | A84S/V85S/S86T/ Q88N/T89P | 8.72 | NB | [41] | ||
| TNFR2 | D143N-A145R | D143N/A145R | NB 2 | 13.1 | [42] | |
| Ligand | Specificity | Variants | Mutation Sites | Binding Affinity (nM) | Ref. | |
|---|---|---|---|---|---|---|
| DR4 | DR5 | |||||
| TRAIL | DR4 & DR5 | G131R | G131R | 8.7 ± 1.0 | 7.9 ± 1.3 | [73] |
| TRAIL-Mu3 | aa 114–121 (VRERGPQR) were replaced by RRRRRRRR | − 1 | − | [74] | ||
| DR4 | Apo2L.DR4–8 | Y213W/S215D/Y189A/ Q193S/N199V/K201R | 2.3-fold to WT | NB 2 | [75] | |
| TRAIL.R1-6 | Y189A/Q193S/N199V/ K201R/Y213 W/S215N | − | − | [76] | ||
| D218H | D218H | 12.3 ± 0.6 | 28.6 ± 1.7 | [77] | ||
| D218Y | D218Y | 107 ± 0.4 | 23.3 ± 0.4 | [77] | ||
| rhTRAILDR4 | S159R | 0.37 ± 0.12 | 4.3 ± 0.9 | [78] | ||
| 4C7 | G131R/R149I/S159R/ N199R/K201H/S215D | 0.021 ± 0.01 | 7.21 ± 4.2 | [79] | ||
| rhTRAIL-C3 | G131R/N199R/K201H | − | − | [80] | ||
| DR5 | Apo2L.DR5–8 | Y189Q/R191K/Q193R/ H264R/I266L/D267Q | NB | 0.8-fold to WT | [75] | |
| TRAIL.R2-6 | Y189Q/R191K/Q193R/ H264R/I266L/D267Q | − | − | [76] | ||
| D269H/E195R | D269H/E195R | 2.9 ± 1.7 | 0.012 ± 0.005 | [81] | ||
| DR5-A | Y189N/R191K/Q193R/ H264R/I266L/D267Q/D269H | NB 2 | 0.33 ± 0.005 | [82] | ||
| DR5-B | Y189N/R191K/Q193R/ H264R/I266L/D269H | NB | 0.71 ± 0.013 | [82] | ||
| Ligand | Specificity | Variants | Mutation Sites | Binding Affinity (pM) | Ref. | |
|---|---|---|---|---|---|---|
| RANK | OPG | |||||
| RANKL | RANK | I248D | I248D | − 1 | − | [99] |
| I248K | I248K | 9 ± 2 | − | [100] | ||
| I248Y | I248Y | 8 ± 3 | − | [100] | ||
| rRANKL5 | aa 246–318 deletion | − | − | [103] | ||
| Q236D | Q236D | 15.0 ± 3.2 | 112.3 ± 24.4 | [104] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suo, F.; Zhou, X.; Setroikromo, R.; Quax, W.J. Receptor Specificity Engineering of TNF Superfamily Ligands. Pharmaceutics 2022, 14, 181. https://doi.org/10.3390/pharmaceutics14010181
Suo F, Zhou X, Setroikromo R, Quax WJ. Receptor Specificity Engineering of TNF Superfamily Ligands. Pharmaceutics. 2022; 14(1):181. https://doi.org/10.3390/pharmaceutics14010181
Chicago/Turabian StyleSuo, Fengzhi, Xinyu Zhou, Rita Setroikromo, and Wim J. Quax. 2022. "Receptor Specificity Engineering of TNF Superfamily Ligands" Pharmaceutics 14, no. 1: 181. https://doi.org/10.3390/pharmaceutics14010181