Inhibitory Effects of Gold and Silver Nanoparticles on the Differentiation into Osteoclasts In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation and Characterization of the GNPs and SNPs
2.3. Isolation of Bone Marrow-Derived Macrophages (BMM) and Primary Culture
2.4. Cytotoxicity Analyze
2.5. TRAP Assay and Measurements
2.6. Actin Ring Formation and Measurements
2.7. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.8. Western Blot
2.9. GNPs and SNPs Uptake Analysis Using Dark Field
2.10. Statistical Analyses
3. Results
3.1. Characterization of GNPs and SNPs
3.2. Cell Viability by GNPs and SNPs in BMMs
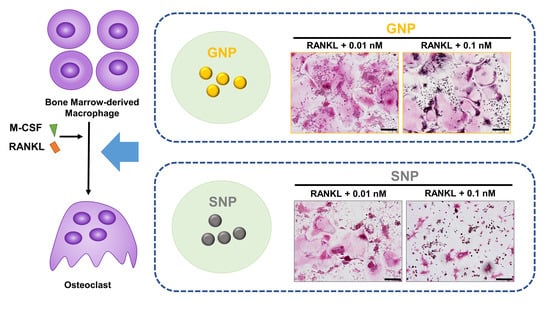
3.3. GNPs and SNPs Inhibited the Formation of TRAP+ MNCs and Actin Rings
3.4. Inhibited Gene Expression by GNPs and SNPs in the RANKL-Induced Osteoclasts
3.5. Effects of GNPs and SNPs in RANKL-Induced NF-κB Signal Pathways
3.6. Comparison of the GNPs and SNPs Uptake Ratio in the BMMs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, Y.; You, X.; Xing, W.; Zhang, Z.; Zou, W. Paracrine and endocrine actions of bone-the functions of secretory proteins from osteoblasts, osteocytes, and osteoclasts. Bone Res. 2018, 6, 1–11. [Google Scholar] [CrossRef]
- Wang, H.H.; Hsu, Y.H.; Chang, M.S. IL-20 bone diseases involvement and therapeutic target potential. J. Biomed. Sci. 2018, 25, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zupan, J.; Komadina, R.; Marc, J. The relationship between osteoclastogenic and anti-osteoclastogenic pro-inflammatory cytokines differs in human osteoporotic and osteoarthritic bone tissues. J. Biomed. Sci. 2012, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Chen, X.; Gao, S.; Yu, X.; Xiao, J.; Zhang, B.; Liu, X.; Dai, M. Key Triggers of Osteoclast-Related Diseases and Available Strategies for Targeted Therapies: A Review. Front. Med. 2017, 4, 234. [Google Scholar] [CrossRef] [PubMed]
- Vial, S.; Reis, R.L.; Oliveira, J.M. Recent advances using gold nanoparticles as a promising multimodal tool for tissue engineering and regenerative medicine. Curr. Opin. Solid State Mater. Sci. 2017, 21, 92–112. [Google Scholar] [CrossRef] [Green Version]
- Tautzenberger, A.; Kovtun, A.; Ignatius, A. Nanoparticles and their potential for application in bone. Int. J. Nanomed. 2012, 7, 4545. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, H.; Nakara, A.; Shanmugam, V.K. Anti-inflammatory mechanism of various metal and metal oxide nanoparticles synthesized using plant extracts: A review. Biomed. Pharmacother. 2019, 109, 2561–2572. [Google Scholar] [CrossRef]
- Ko, W.K.; Kim, S.J.; Heo, D.N.; Han, I.B.; Kim, S.; Kwon, I.K.; Sohn, S. Double layers of gold nanoparticles immobilized titanium implants improve the osseointegration in rabbit models. Nanomedicine 2020, 24, 102129. [Google Scholar] [CrossRef]
- Qin, H.; Zhu, C.; An, Z.; Jiang, Y.; Zhao, Y.; Wang, J.; Liu, X.; Hui, B.; Zhang, X.; Wang, Y. Silver nanoparticles promote osteogenic differentiation of human urine-derived stem cells at noncytotoxic concentrations. Int. J. Nanomed. 2014, 9, 2469–2478. [Google Scholar] [CrossRef] [Green Version]
- Abdal Dayem, A.; Lee, S.B.; Cho, S.-G. The impact of metallic nanoparticles on stem cell proliferation and differentiation. Nanomaterials 2018, 8, 761. [Google Scholar] [CrossRef] [Green Version]
- Castiglioni, S.; Cazzaniga, A.; Locatelli, L.; Maier, J.A. Silver nanoparticles in orthopedic applications: New insights on their effects on osteogenic cells. Nanomaterials 2017, 7, 124. [Google Scholar] [CrossRef] [Green Version]
- Sul, O.-J.; Kim, J.-C.; Kyung, T.-W.; Kim, H.-J.; Kim, Y.-Y.; Kim, S.-H.; Kim, J.-S.; Choi, H.-S. Gold nanoparticles inhibited the receptor activator of nuclear factor-κb ligand (RANKL)-induced osteoclast formation by acting as an antioxidant. Biosci. Biotechnol. Biochem. 2010, 74, 2209–2213. [Google Scholar] [CrossRef] [PubMed]
- Nah, H.; Lee, D.; Lee, J.S.; Lee, S.J.; Heo, D.N.; Lee, Y.-H.; Bang, J.B.; Hwang, Y.-S.; Moon, H.-J.; Kwon, I.K. Strategy to inhibit effective differentiation of RANKL-induced osteoclasts using vitamin D-conjugated gold nanoparticles. Appl. Surf. Sci. 2020, 527, 146765. [Google Scholar] [CrossRef]
- Heo, D.N.; Ko, W.-K.; Moon, H.-J.; Kim, H.-J.; Lee, S.J.; Lee, J.B.; Bae, M.S.; Yi, J.-K.; Hwang, Y.-S.; Bang, J.B. Inhibition of osteoclast differentiation by gold nanoparticles functionalized with cyclodextrin curcumin complexes. ACS Nano 2014, 8, 12049–12062. [Google Scholar] [CrossRef]
- Lee, D.; Heo, D.N.; Kim, H.J.; Ko, W.K.; Lee, S.J.; Heo, M.; Bang, J.B.; Lee, J.B.; Hwang, D.S.; Do, S.H.; et al. Inhibition of Osteoclast Differentiation and Bone Resorption by Bisphosphonate-conjugated Gold Nanoparticles. Sci. Rep. 2016, 6, 27336. [Google Scholar] [CrossRef]
- Hughes, H.K. Beer’s law and the optimum transmittance in absorption measurements. Appl. Opt. 1963, 2, 937–945. [Google Scholar] [CrossRef]
- Paramelle, D.; Sadovoy, A.; Gorelik, S.; Free, P.; Hobley, J.; Fernig, D.G. A rapid method to estimate the concentration of citrate capped silver nanoparticles from UV-visible light spectra. Analyst 2014, 139, 4855–4861. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Ko, W.-K.; Jo, M.-J.; Arai, Y.; Choi, H.; Kumar, H.; Han, I.-B.; Sohn, S. Anti-inflammatory effect of tauroursodeoxycholic acid in RAW 264.7 macrophages, bone marrow-derived macrophages, BV2 microglial cells, and spinal cord injury. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Han, Y.; Xie, F.; Ma, X.; Xu, Z.; Liu, X.; Zou, W.; Wang, J. A RANKL-based Osteoclast Culture Assay of Mouse Bone Marrow to Investigate the Role of mTORC1 in Osteoclast Formation. J. Vis. Exp. 2018, 133, 56468. [Google Scholar] [CrossRef]
- Ko, W.K.; Lee, S.H.; Kim, S.J.; Jo, M.J.; Kumar, H.; Han, I.B.; Sohn, S. Anti-inflammatory effects of ursodeoxycholic acid by lipopolysaccharide-stimulated inflammatory responses in RAW 264.7 macrophages. PLoS ONE 2017, 12, e0180673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishimoto, Y.; Fukumoto, S.; Nishihara, S.; Mizumura, H.; Hirai, K.; Teshima, R. Gene expression relevant to osteoclastogenesis in the synovium and bone marrow of mature rats with collagen-induced arthritis. Rheumatology 2004, 43, 1496–1503. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Ding, S.; Yin, D.; Cuan, X.; Xie, C.; Xu, H.; Wang, X.; Sheng, J. Pu-erh Tea Extract Ameliorates Ovariectomy-Induced Osteoporosis in Rats and Suppresses Osteoclastogenesis In Vitro. Front. Pharmacol. 2017, 8, 324. [Google Scholar] [CrossRef] [Green Version]
- Matsubara, R.; Kukita, T.; Ichigi, Y.; Takigawa, I.; Qu, P.F.; Funakubo, N.; Miyamoto, H.; Nonaka, K.; Kukita, A. Characterization and identification of subpopulations of mononuclear preosteoclasts induced by TNF-alpha in combination with TGF-beta in rats. PLoS ONE 2012, 7, e47930. [Google Scholar] [CrossRef] [Green Version]
- Han, G.H.; Kim, S.J.; Ko, W.-K.; Lee, D.; Lee, J.S.; Nah, H.; Han, I.-B.; Sohn, S. Injectable Hydrogel Containing Tauroursodeoxycholic Acid for Anti-neuroinflammatory Therapy After Spinal Cord Injury in Rats. Mol. Neurobiol. 2020, 57, 4007–4017. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Ko, W.-K.; Heo, D.N.; Lee, S.J.; Lee, D.; Heo, M.; Han, I.-B.; Kwon, I.K.; Sohn, S. Anti-neuroinflammatory gold nanocomplex loading ursodeoxycholic acid following spinal cord injury. Chem. Eng. J. 2019, 375, 122088. [Google Scholar] [CrossRef]
- He, Y.Q.; Liu, S.P.; Kong, L.; Liu, Z.F. A study on the sizes and concentrations of gold nanoparticles by spectra of absorption, resonance Rayleigh scattering and resonance non-linear scattering. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2005, 61, 2861–2866. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Kim, N. Signaling pathways in osteoclast differentiation. Chonnam Med. J. 2016, 52, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orimo, H.; Hayashi, Y.; Fukunaga, M.; Sone, T.; Fujiwara, S.; Shiraki, M.; Kushida, K.; Miyamoto, S.; Soen, S.; Nishimura, J. Diagnostic criteria for primary osteoporosis: Year 2000 revision. J. Bone Miner. Metab. 2001, 19, 331. [Google Scholar] [CrossRef] [PubMed]
- Ko, W.K.; Heo, D.N.; Moon, H.J.; Lee, S.J.; Bae, M.S.; Lee, J.B.; Sun, I.C.; Jeon, H.B.; Park, H.K.; Kwon, I.K. The effect of gold nanoparticle size on osteogenic differentiation of adipose-derived stem cells. J. Colloid Interface Sci. 2015, 438, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Jin, C.; Ma, L.; Feng, X.; Deng, X.; Wu, S.; Liu, X.; Yang, C. Accelerated Bone Regeneration by Gold-Nanoparticle-Loaded Mesoporous Silica through Stimulating Immunomodulation. ACS Appl. Mater. Interfaces 2019, 11, 41758–41769. [Google Scholar] [CrossRef]
- Albers, C.E.; Hofstetter, W.; Siebenrock, K.A.; Landmann, R.; Klenke, F.M. In vitro cytotoxicity of silver nanoparticles on osteoblasts and osteoclasts at antibacterial concentrations. Nanotoxicology 2013, 7, 30–36. [Google Scholar] [CrossRef]
- Lee, S.J.; Heo, D.N.; Moon, J.H.; Ko, W.K.; Lee, J.B.; Bae, M.S.; Park, S.W.; Kim, J.E.; Lee, D.H.; Kim, E.C.; et al. Electrospun chitosan nanofibers with controlled levels of silver nanoparticles. Preparation, characterization and antibacterial activity. Carbohydr. Polym. 2014, 111, 530–537. [Google Scholar] [CrossRef]
- De Giglio, E.; Cafagna, D.; Cometa, S.; Allegretta, A.; Pedico, A.; Giannossa, L.; Sabbatini, L.; Mattioli-Belmonte, M.; Iatta, R. An innovative, easily fabricated, silver nanoparticle-based titanium implant coating: Development and analytical characterization. Anal. Bioanal. Chem. 2013, 405, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.; Lee, S.J.; Moon, J.-H.; Kim, J.H.; Heo, D.N.; Bang, J.B.; Lim, H.-N.; Kwon, I.K. Preparation of antibacterial chitosan membranes containing silver nanoparticles for dental barrier membrane applications. J. Ind. Eng. Chem. 2018, 66, 196–202. [Google Scholar] [CrossRef]
- Kirmanidou, Y.; Sidira, M.; Bakopoulou, A.; Tsouknidas, A.; Prymak, O.; Papi, R.; Choli-Papadopoulou, T.; Epple, M.; Michailidis, N.; Koidis, P. Assessment of cytotoxicity and antibacterial effects of silver nanoparticle-doped titanium alloy surfaces. Dent. Mater. 2019, 35, e220–e233. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Kienzle, A.; Liu, X.; Müller, W.E.; Elkhooly, T.A.; Feng, Q. In vitro effect of 30 nm silver nanoparticles on adipogenic differentiation of human mesenchymal stem cells. J. Biomed. Nanotechnol. 2016, 12, 525–535. [Google Scholar] [CrossRef]
- Edwards, J.R.; Mundy, G.R. Advances in osteoclast biology: Old findings and new insights from mouse models. Nat. Rev. Rheumatol. 2011, 7, 235. [Google Scholar] [CrossRef]
- Matsubara, T.; Kinbara, M.; Maeda, T.; Yoshizawa, M.; Kokabu, S.; Takano Yamamoto, T. Regulation of osteoclast differentiation and actin ring formation by the cytolinker protein plectin. Biochem. Biophys. Res. Commun. 2017, 489, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Itoh, H.; Kawazoe, Y.; Miyazaki, S.; Doi, K.; Kubo, T.; Akagawa, Y.; Shiba, T. Polyphosphate-mediated inhibition of tartrate-resistant acid phosphatase and suppression of bone resorption of osteoclasts. PLoS ONE 2013, 8, e78612. [Google Scholar] [CrossRef] [Green Version]
- Lotinun, S.; Kiviranta, R.; Matsubara, T.; Alzate, J.A.; Neff, L.; Lüth, A.; Koskivirta, I.; Kleuser, B.; Vacher, J.; Vuorio, E. Osteoclast-specific cathepsin K deletion stimulates S1P-dependent bone formation. J. Clin. Investig. 2013, 123. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Mei, J.; Shao, D.; Zhou, F.; Qiao, H.; Liang, Y.; Li, K.; Tang, T. Cerium Oxide Nanoparticles Regulate Osteoclast Differentiation Bidirectionally by Modulating the Cellular Production of Reactive Oxygen Species. Int. J. Nanomed. 2020, 15, 6355. [Google Scholar] [CrossRef] [PubMed]
- Yeung, T.; Gilbert, G.E.; Shi, J.; Silvius, J.; Kapus, A.; Grinstein, S. Membrane phosphatidylserine regulates surface charge and protein localization. Science 2008, 319, 210–213. [Google Scholar] [CrossRef]
- Zhao, F.; Zhao, Y.; Liu, Y.; Chang, X.; Chen, C.; Zhao, Y. Cellular uptake, intracellular trafficking, and cytotoxicity of nanomaterials. Small 2011, 7, 1322–1337. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.-G.; Wei, W.; Lv, P.-P.; Yue, H.; Wang, L.-Y.; Su, Z.-G.; Ma, G.-H. Surface charge affects cellular uptake and intracellular trafficking of chitosan-based nanoparticles. Biomacromolecules 2011, 12, 2440–2446. [Google Scholar] [CrossRef] [PubMed]
- Fontana, F.; Figueiredo, P.; Bauleth-Ramos, T.; Correia, A.; Santos, H.A. Immunostimulation and immunosuppression: Nanotechnology on the brink. Small Methods 2018, 2, 1700347. [Google Scholar] [CrossRef]
- Krishnaraj, C.; Harper, S.L.; Yun, S.-I. In Vivo toxicological assessment of biologically synthesized silver nanoparticles in adult Zebrafish (Danio rerio). J. Hazardous Mater. 2016, 301, 480–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.J.; Rehman, S.U.; Amin, F.U.; Kim, M.O. Enhanced neuroprotection of anthocyanin-loaded PEG-gold nanoparticles against Aβ1-42-induced neuroinflammation and neurodegeneration via the NF-KB/JNK/GSK3β signaling pathway. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2533–2544. [Google Scholar] [CrossRef]
- David, L.; Moldovan, B.; Baldea, I.; Olteanu, D.; Bolfa, P.; Clichici, S.; Filip, G.A. Modulatory effects of Cornus sanguinea L. mediated green synthesized silver nanoparticles on oxidative stress, COX-2/NOS2 and NFkB/pNFkB expressions in experimental inflammation in Wistar rats. Mater. Sci. Eng. C 2020, 110, 110709. [Google Scholar] [CrossRef]
- Bai, X.; Gao, Y.; Zhang, M.; Chang, Y.-N.; Chen, K.; Li, J.; Zhang, J.; Liang, Y.; Kong, J.; Wang, Y. Carboxylated gold nanoparticles inhibit bone erosion by disturbing the acidification of an osteoclast absorption microenvironment. Nanoscale 2020, 12, 3871–3878. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, D.; Ko, W.-K.; Kim, S.J.; Han, I.-B.; Hong, J.B.; Sheen, S.H.; Sohn, S. Inhibitory Effects of Gold and Silver Nanoparticles on the Differentiation into Osteoclasts In Vitro. Pharmaceutics 2021, 13, 462. https://doi.org/10.3390/pharmaceutics13040462
Lee D, Ko W-K, Kim SJ, Han I-B, Hong JB, Sheen SH, Sohn S. Inhibitory Effects of Gold and Silver Nanoparticles on the Differentiation into Osteoclasts In Vitro. Pharmaceutics. 2021; 13(4):462. https://doi.org/10.3390/pharmaceutics13040462
Chicago/Turabian StyleLee, Daye, Wan-Kyu Ko, Seong Jun Kim, In-Bo Han, Je Beom Hong, Seung Hun Sheen, and Seil Sohn. 2021. "Inhibitory Effects of Gold and Silver Nanoparticles on the Differentiation into Osteoclasts In Vitro" Pharmaceutics 13, no. 4: 462. https://doi.org/10.3390/pharmaceutics13040462