Aliphatic Quaternary Ammonium Functionalized Nanogels for Gene Delivery
Abstract
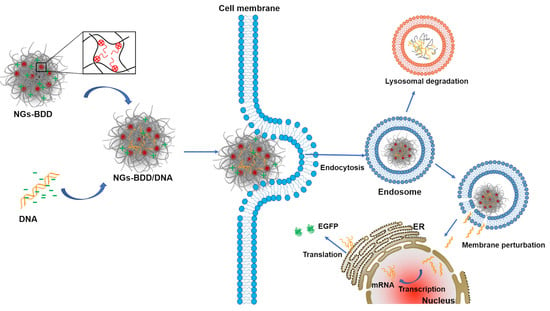
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Instruments and Methods
2.3. Synthesis of the Nanogels
2.3.1. Synthesis of the Non-Quaternized Nanogel (NGs)
2.3.2. Quaternization of Nanogel with Methyl Iodide (NGs-MI)
2.3.3. Quaternization of Nanogel with 1-Bromododecane (NGs-BDD)
2.4. Buffering Capacities
2.5. Gel Retardation Electrophoresis
2.6. Cytotoxicity Assay
2.7. In Vitro Transfection
2.8. Cellular Uptake and Colocalization with Lysotracker
2.9. Endosomal Escape of ODNs Mediated by Nanogel/ODNs Complexes
2.10. Membrane Perturbing Activity of Nanogels and Nanogel/pDNA Complexes
2.11. Statistics
3. Results and Discussion
3.1. Preparation and Characterization of Nanogels
3.2. Complexes of Nanogels and Plasmid DNA
3.3. Cellular Uptake and Endosomal Escape of Nanogel/pDNA Complexes
3.4. In Vitro Transfection with NGs-BDD/pDNA Complexes
3.5. Membrane Perturbing Activity Is Responsible for NGs-BDD Mediated Transfection Capacity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, Z.H.; Wang, J.; Gu, Z.L. Development of Novel Therapies for Huntington’s Disease: Hope and Challenge. Acta Pharmacol. Sin. 2005, 26, 129–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendell, J.R.; Campbell, K.; Rodino-Klapac, L.; Sahenk, Z.; Shilling, C.; Lewis, S.; Bowles, D.; Gray, S.; Li, C.; Galloway, G.; et al. Dystrophin Immunity in Duchenne’s Muscular Dystrophy. N. Engl. J. Med. 2010, 363, 1429–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplitt, M.G.; Feigin, A.; Tang, C.; Fitzsimons, H.L.; Mattis, P.; Lawlor, P.A.; Bland, R.J.; Young, D.; Strybing, K.; Eidelberg, D.; et al. Safety and Tolerability of Gene Therapy with an Adeno-Associated Virus (AAV) Borne GAD Gene for Parkinson’s Disease: An Open Label, Phase I Trial. Lancet 2007, 369, 2097–2105. [Google Scholar] [CrossRef]
- Ozpolat, B.; Sood, A.K.; Lopez-Berestein, G. Liposomal SiRNA Nanocarriers for Cancer Therapy. Adv. Drug Deliv. Rev. 2014, 66, 110–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Couzin, J. Gene Therapy: As Gelsinger Case Ends, Gene Therapy Suffers Another Blow. Science 2005, 307, 1028b. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Brown, A.M.; Jenkins, C.; Campbell, K. Viral Vector Systems for Gene Therapy: A Comprehensive Literature Review of Progress and Biosafety Challenges. Appl. Biosaf. 2020, 25, 7–18. [Google Scholar] [CrossRef]
- Mastrobattista, E.; Hennink, W.E. Charged for Success. Nat. Mater. 2012, 11, 10–12. [Google Scholar] [CrossRef]
- Salameh, J.W.; Zhou, L.; Ward, S.M.; Santa Chalarca, C.F.; Emrick, T.; Figueiredo, M.L. Polymer-mediated Gene Therapy: Recent Advances and Merging of Delivery Techniques. WIREs Nanomed. Nanobiotechnol. 2020, 12, e1598. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Cudjoe, D.K.; Song, W. Multicomponent Polymerization toward Cationic Polymers for Efficient Gene Delivery. Macromol. Rapid Commun. 2021, 42, 2000464. [Google Scholar] [CrossRef]
- Alabi, C.A.; Love, K.T.; Sahay, G.; Yin, H.; Luly, K.M.; Langer, R.; Anderson, D.G. Multiparametric Approach for the Evaluation of Lipid Nanoparticles for SiRNA Delivery. Proc. Natl. Acad. Sci. USA 2013, 110, 12881–12886. [Google Scholar] [CrossRef] [Green Version]
- Buck, J.; Grossen, P.; Cullis, P.R.; Huwyler, J.; Witzigmann, D. Lipid-Based DNA Therapeutics: Hallmarks of Non-Viral Gene Delivery. ACS Nano 2019, 13, 3754–3782. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Qin, B.; Yin, H.; Shi, Y.; Jiang, M.; Luo, L.; Luo, Z.; Zhang, J.; Li, X.; Zhu, C.; et al. Virus-like Nonvirus Cationic Liposome for Efficient Gene Delivery via Endoplasmic Reticulum Pathway. ACS Cent. Sci. 2020, 6, 174–188. [Google Scholar] [CrossRef]
- Barba, A.A.; Bochicchio, S.; Dalmoro, A.; Lamberti, G. Lipid Delivery Systems for Nucleic-Acid-Based-Drugs: From Production to Clinical Applications. Pharmaceutics 2019, 11, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, C.J.; Kozielski, K.L.; Green, J.J. Exploring the Role of Polymer Structure on Intracellular Nucleic Acid Delivery via Polymeric Nanoparticles. J. Control. Release 2015, 219, 488–499. [Google Scholar] [CrossRef] [Green Version]
- Fayazpour, F.; Lucas, B.; Alvarez-Lorenzo, C.; Sanders, N.N.; Demeester, J.; De Smedt, S.C. Physicochemical and Transfection Properties of Cationic Hydroxyethylcellulose/DNA Nanoparticles. Biomacromolecules 2006, 7, 2856–2862. [Google Scholar] [CrossRef] [PubMed]
- Yudovin-Farber, I.; Yanay, C.; Azzam, T.; Linial, M.; Domb, A.J. Quaternary Ammonium Polysaccharides for Gene Delivery. Bioconjug. Chem. 2005, 16, 1196–1203. [Google Scholar] [CrossRef]
- Boussif, O.; LezoualC’H, F.; Zanta, M.A.; Mergny, M.D.; Scherman, D.; Demeneix, B.; Behr, J.P. A Versatile Vector for Gene and Oligonucleotide Transfer into Cells in Culture and in Vivo: Polyethylenimine. Proc. Natl. Acad. Sci. USA 1995, 92, 7297–7301. [Google Scholar] [CrossRef] [Green Version]
- Rehman, Z.U.; Hoekstra, D.; Zuhorn, I.S. Mechanism of Polyplex- and Lipoplex-Mediated Delivery of Nucleic Acids: Real-Time Visualization of Transient Membrane Destabilization without Endosomal Lysis. ACS Nano 2013, 7, 3767–3777. [Google Scholar] [CrossRef]
- Degors, I.M.S.; Wang, C.; Rehman, Z.U.; Zuhorn, I.S. Carriers Break Barriers in Drug Delivery: Endocytosis and Endosomal Escape of Gene Delivery Vectors. Acc. Chem. Res. 2019, 52, 1750–1760. [Google Scholar] [CrossRef] [Green Version]
- Zuhorn, I.S.; Bakowsky, U.; Polushkin, E.; Visser, W.H.; Stuart, M.C.A.; Engberts, J.B.F.N.; Hoekstra, D. Nonbilayer Phase of Lipoplex-Membrane Mixture Determines Endosomal Escape of Genetic Cargo and Transfection Efficiency. Mol. Ther. 2005, 11, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Koltover, I. An Inverted Hexagonal Phase of Cationic Liposome-DNA Complexes Related to DNA Release and Delivery. Science 1998, 281, 78–81. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, J.A.; Cullis, P.R.; Van Der Meel, R. Lipid Nanoparticles Enabling Gene Therapies: From Concepts to Clinical Utility. Nucleic Acid Ther. 2018, 28, 146–157. [Google Scholar] [CrossRef] [Green Version]
- Das, M.; Mardyani, S.; Chan, W.C.W.; Kumacheva, E. Biofunctionalized PH-Responsive Microgels for Cancer Cell Targeting: Rational Design. Adv. Mater. 2006, 18, 80–83. [Google Scholar] [CrossRef]
- Vinogradov, S. Colloidal Microgels in Drug Delivery Applications. Curr. Pharm. Des. 2006, 12, 4703–4712. [Google Scholar] [CrossRef] [Green Version]
- Nayak, S.; Lee, H.; Chmielewski, J.; Lyon, L.A. Folate-Mediated Cell Targeting and Cytotoxicity Using Thermoresponsive Microgels. J. Am. Chem. Soc. 2004, 126, 10258–10259. [Google Scholar] [CrossRef]
- Blackburn, W.H.; Lyon, L.A. Size-Controlled Synthesis of Monodisperse Core/Shell Nanogels. Colloid Polym. Sci. 2008, 286, 563–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malonne, H.; Eeckman, F.; Fontaine, D.; Otto, A.; De Vos, L.; Moës, A.; Fontaine, J.; Amighi, K. Preparation of Poly(N-Isopropylacrylamide) Copolymers and Preliminary Assessment of Their Acute and Subacute Toxicity in Mice. Eur. J. Pharm. Biopharm. 2005, 61, 188–194. [Google Scholar] [CrossRef]
- Shin, Y.; Chang, J.H.; Liu, J.; Williford, R.; Shin, Y.K.; Exarhos, G.J. Hybrid Nanogels for Sustainable Positive Thermosensitive Drug Release. J. Control. Release 2001, 73, 1–6. [Google Scholar] [CrossRef]
- Plamper, F.A.; Richtering, W. Functional Microgels and Microgel Systems. Acc. Chem. Res. 2017, 50, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.; Imaz, A.; Forcada, J. Temperature-Sensitive Nanogels: Poly(N-Vinylcaprolactam) versus Poly(N-Isopropylacrylamide). Polym. Chem. 2012, 3, 852–856. [Google Scholar] [CrossRef]
- Yang, H.; Wang, Q.; Huang, S.; Xiao, A.; Li, F.; Gan, L.; Yang, X. Smart PH/Redox Dual-Responsive Nanogels for On-Demand Intracellular Anticancer Drug Release. ACS Appl. Mater. Interfaces 2016, 8, 7729–7738. [Google Scholar] [CrossRef] [PubMed]
- Pelton, R. Temperature-Sensitive Aqueous Microgels. Adv. Colloid Interface Sci. 2000, 85, 1–33. [Google Scholar] [CrossRef]
- Tan, K.H.; Xu, W.; Stefka, S.; Demco, D.E.; Kharandiuk, T.; Ivasiv, V.; Nebesnyi, R.; Petrovskii, V.S.; Potemkin, I.I.; Pich, A. Selenium-Modified Microgels as Bio-Inspired Oxidation Catalysts. Angew. Chem. Int. Ed. 2019, 58, 9791–9796. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, G.; Schürings, M.P.; Van Rijn, P.; Pich, A. Formation of Catalytically Active Gold-Polymer Microgel Hybrids via a Controlled in Situ Reductive Process. J. Mater. Chem. A 2013, 1, 13244–13251. [Google Scholar] [CrossRef]
- Das, S.S.; Bharadwaj, P.; Bilal, M.; Barani, M.; Rahdar, A.; Taboada, P.; Bungau, S.; Kyzas, G.Z. Stimuli-Responsive Polymeric Nanocarriers for Drug Delivery, Imaging, and Theragnosis. Polymers 2020, 12, 1397. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhou, X.; Wang, G. Selective Release of Hydrophobic and Hydrophilic Cargos from Multi-Stimuli-Responsive Nanogels. ACS Appl. Mater. Interfaces 2016, 8, 28888–28896. [Google Scholar] [CrossRef]
- Meurer, R.A.; Kemper, S.; Knopp, S.; Eichert, T.; Jakob, F.; Goldbach, H.E.; Schwaneberg, U.; Pich, A. Biofunctional Microgel-Based Fertilizers for Controlled Foliar Delivery of Nutrients to Plants. Angew. Chem. Int. Ed. 2017, 56, 7380–7386. [Google Scholar] [CrossRef]
- Keskin, D.; Mergel, O.; Van Der Mei, H.C.; Busscher, H.J.; Van Rijn, P. Inhibiting Bacterial Adhesion by Mechanically Modulated Microgel Coatings. Biomacromolecules 2019, 20, 243–253. [Google Scholar] [CrossRef] [Green Version]
- Keskin, D.; Zu, G.; Forson, A.M.; Tromp, L.; Sjollema, J.; van Rijn, P. Nanogels: A Novel Approach in Antimicrobial Delivery Systems and Antimicrobial Coatings. Bioact. Mater. 2021, 6, 3634–3657. [Google Scholar] [CrossRef]
- Guan, Y.; Zhang, Y. PNIPAM Microgels for Biomedical Applications: From Dispersed Particles to 3D Assemblies. Soft Matter 2011, 7, 6375. [Google Scholar] [CrossRef]
- Blackburn, W.H.; Dickerson, E.B.; Smith, M.H.; McDonald, J.F.; Lyon, L.A. Peptide-Functionalized Nanogels for Targeted SiRNA Delivery. Bioconjug. Chem. 2009, 20, 960–968. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, S.; Patil, S.; Singh, N. Enhancing Gene-Knockdown Efficiency of Poly(N-Isopropylacrylamide) Nanogels. ACS Omega 2018, 3, 8042–8049. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.H.; Lyon, L.A. Multifunctional Nanogels for SiRNA Delivery. Acc. Chem. Res. 2012, 45, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Ribovski, L.; de Jong, E.; Mergel, O.; Zu, G.; Keskin, D.; van Rijn, P.; Zuhorn, I.S. Low Nanogel Stiffness Favors Nanogel Transcytosis across an in Vitro Blood–Brain Barrier. Nanomed. Nanotechnol. Biol. Med. 2021, 34, 102377. [Google Scholar] [CrossRef]
- Zu, G.; Steinmüller, M.; Keskin, D.; Van Der Mei, H.C.; Mergel, O.; Van Rijn, P. Antimicrobial Nanogels with Nanoinjection Capabilities for Delivery of the Hydrophobic Antibacterial Agent Triclosan. ACS Appl. Polym. Mater. 2020, 2, 5779–5789. [Google Scholar] [CrossRef]
- Keskin, D.; Tromp, L.; Mergel, O.; Zu, G.; Warszawik, E.; Van Der Mei, H.C.; Van Rijn, P. Highly Efficient Antimicrobial and Antifouling Surface Coatings with Triclosan-Loaded Nanogels. ACS Appl. Mater. Interfaces 2020, 12, 57721–57731. [Google Scholar] [CrossRef] [PubMed]
- Mergel, O.; Gelissen, A.P.H.; Wünnemann, P.; Böker, A.; Simon, U.; Plamper, F.A. Selective Packaging of Ferricyanide within Thermoresponsive Microgels. J. Phys. Chem. C 2014, 118, 26199–26211. [Google Scholar] [CrossRef]
- Yue, J.; Zhao, P.; Gerasimov, J.Y.; Van De Lagemaat, M.; Grotenhuis, A.; Rustema-Abbing, M.; Van Der Mei, H.C.; Busscher, H.J.; Herrmann, A.; Ren, Y. 3D-Printable Antimicrobial Composite Resins. Adv. Funct. Mater. 2015, 25, 6756–6767. [Google Scholar] [CrossRef]
- Zuhorn, I.S.; Oberle, V.; Visser, W.H.; Engberts, J.B.F.N.; Bakowsky, U.; Polushkin, E.; Hoekstra, D. Phase Behavior of Cationic Amphiphiles and Their Mixtures with Helper Lipid Influences Lipoplex Shape, DNA Translocation, and Transfection Efficiency. Biophys. J. 2002, 83, 2096–2108. [Google Scholar] [CrossRef] [Green Version]
- Sigolaeva, L.; Pergushov, D.; Oelmann, M.; Schwarz, S.; Brugnoni, M.; Kurochkin, I.; Plamper, F.; Fery, A.; Richtering, W. Surface Functionalization by Stimuli-Sensitive Microgels for Effective Enzyme Uptake and Rational Design of Biosensor Setups. Polymers 2018, 10, 791. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Urban, B.; Reis, B.; Yu, X.; Grab, A.; Cavalcanti-Adam, E.; Kuckling, D. Switchable Release of Bone Morphogenetic Protein from Thermoresponsive Poly(NIPAM-co-DMAEMA)/Cellulose Sulfate Particle Coatings. Polymers 2018, 10, 1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Keskin, D.; de Haan-Visser, W.H.; Zu, G.; van Rijn, P.; Zuhorn, I.S. Aliphatic Quaternary Ammonium Functionalized Nanogels for Gene Delivery. Pharmaceutics 2021, 13, 1964. https://doi.org/10.3390/pharmaceutics13111964
Zhang H, Keskin D, de Haan-Visser WH, Zu G, van Rijn P, Zuhorn IS. Aliphatic Quaternary Ammonium Functionalized Nanogels for Gene Delivery. Pharmaceutics. 2021; 13(11):1964. https://doi.org/10.3390/pharmaceutics13111964
Chicago/Turabian StyleZhang, Huaiying, Damla Keskin, Willy H. de Haan-Visser, Guangyue Zu, Patrick van Rijn, and Inge S. Zuhorn. 2021. "Aliphatic Quaternary Ammonium Functionalized Nanogels for Gene Delivery" Pharmaceutics 13, no. 11: 1964. https://doi.org/10.3390/pharmaceutics13111964