miRNA Reference Genes in Extracellular Vesicles Released from Amniotic Membrane-Derived Mesenchymal Stromal Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
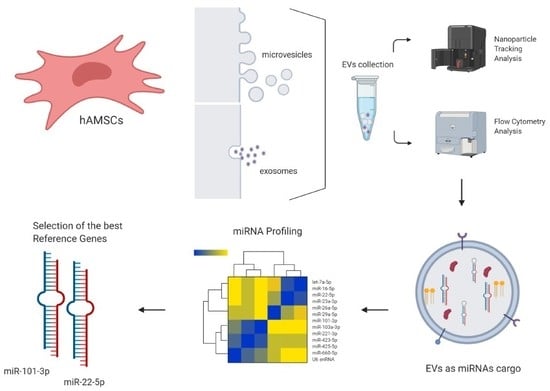
2.2. hAMSC Isolation and Expansion
2.3. hAMSC Characterization
2.4. hAMSC-EV Isolation and Characterization
2.5. Candidate RG Selection
2.6. Total RNA Isolation and miRNA Profiling
2.7. Data Analysis
2.8. Statistical Analysis
3. Results
3.1. hAMSCs and EVs Characterization
3.2. Expression of Candidate Reference Genes
3.3. RGs Stability Analysis
3.4. Impact of RGs Choice on the Quantification of Target Genes
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yin, K.; Wang, S.; Zhao, R.C. Exosomes from mesenchymal stem/stromal cells: A new therapeutic paradigm. Biomark. Res. 2019, 7, 8. [Google Scholar] [CrossRef] [Green Version]
- Deng, H.; Sun, C.; Sun, Y.; Li, H.; Yang, L.; Wu, D.; Gao, Q.; Jiang, X. Lipid, Protein, and MicroRNA Composition Within Mesenchymal Stem Cell-Derived Exosomes. Cell Reprogram 2018, 20, 178–186. [Google Scholar] [CrossRef]
- Qiu, G.; Zheng, G.; Ge, M.; Wang, J.; Huang, R.; Shu, Q.; Xu, J. Mesenchymal stem cell-derived extracellular vesicles affect disease outcomes via transfer of microRNAs. Stem Cell Res. Ther. 2018, 9, 320. [Google Scholar] [CrossRef]
- Naseri, Z.; Oskuee, R.K.; Jaafari, M.R.; Forouzandeh Moghadam, M. Exosome-mediated delivery of functionally active miRNA-142-3p inhibitor reduces tumorigenicity of breast cancer in vitro and in vivo. Int. J. Nanomed. 2018, 13, 7727–7747. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.C.; Yuan, T.; Zhang, Y.L.; Yin, W.J.; Guo, S.C.; Zhang, C.Q. Exosomes derived from miR-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics 2017, 7, 180–195. [Google Scholar] [CrossRef]
- Barter, M.J.; Tselepi, M.; Gómez, R.; Woods, S.; Hui, W.; Smith, G.R.; Shanley, D.P.; Clark, I.M.; Young, D.A. Genome-Wide MicroRNA and Gene Analysis of Mesenchymal Stem Cell Chondrogenesis Identifies an Essential Role and Multiple Targets for miR-140-5p. Stem Cells 2015, 33, 3266–3280. [Google Scholar] [CrossRef] [Green Version]
- Park, K.S.; Bandeira, E.; Shelke, G.V.; Lässer, C.; Lötvall, J. Enhancement of therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Stem Cell Res. Ther. 2019, 10, 288. [Google Scholar] [CrossRef]
- Ragni, E.; Perucca Orfei, C.; De Luca, P.; Viganò, M.; Colombini, A.; Lugano, G.; Bollati, V.; de Girolamo, L. miR-22-5p and miR-29a-5p Are Reliable Reference Genes for Analyzing Extracellular Vesicle-Associated miRNAs in Adipose-Derived Mesenchymal Stem Cells and Are Stable under Inflammatory Priming Mimicking Osteoarthritis Condition. Stem Cell Rev. Rep. 2019, 15, 743–754. [Google Scholar] [CrossRef]
- Ragni, E.; Perucca Orfei, C.; De Luca, P.; Colombini, A.; Viganò, M.; de Girolamo, L. Secreted Factors and EV-miRNAs Orchestrate the Healing Capacity of Adipose Mesenchymal Stem Cells for the Treatment of Knee Osteoarthritis. Int. J. Mol. Sci. 2020, 21, 1582. [Google Scholar] [CrossRef] [Green Version]
- Parolini, O.; Alviano, F.; Bagnara, G.P.; Bilic, G.; Buhring, H.J.; Evangelista, M.; Hennerbichler, S.; Liu, B.; Magatti, M.; Mao, N.; et al. Concise review: Isolation and characterization of cells from human term placenta: Outcome of the first international Workshop on Placenta Derived Stem Cells. Stem Cells 2008, 26, 300–311. [Google Scholar] [CrossRef] [Green Version]
- Cargnoni, A.; Gibelli, L.; Tosini, A.; Signoroni, P.B.; Nassuato, C.; Arienti, D.; Lombardi, G.; Albertini, A.; Wengler, G.S.; Parolini, O. Transplantation of allogeneic and xenogeneic placenta-derived cells reduces bleomycin-induced lung fibrosis. Cell Transplant. 2009, 18, 405–422. [Google Scholar] [CrossRef] [Green Version]
- Cargnoni, A.; Ressel, L.; Rossi, D.; Poli, A.; Arienti, D.; Lombardi, G.; Parolini, O. Conditioned medium from amniotic mesenchymal tissue cells reduces progression of bleomycin-induced lung fibrosis. Cytotherapy 2012, 14, 153–161. [Google Scholar] [CrossRef] [Green Version]
- Carbone, A.; Castellani, S.; Favia, M.; Diana, A.; Paracchini, V.; Di Gioia, S.; Seia, M.; Casavola, V.; Colombo, C.; Conese, M. Correction of defective CFTR/ENaC function and tightness of cystic fibrosis airway epithelium by amniotic mesenchymal stromal (stem) cells. J. Cell. Mol. Med. 2014, 18, 1631–1643. [Google Scholar] [CrossRef]
- Cargnoni, A.; Piccinelli, E.C.; Ressel, L.; Rossi, D.; Magatti, M.; Toschi, I.; Cesari, V.; Albertini, M.; Mazzola, S.; Parolini, O. Conditioned medium from amniotic membrane-derived cells prevents lung fibrosis and preserves blood gas exchanges in bleomycin-injured mice-specificity of the effects and insights into possible mechanisms. Cytotherapy 2014, 16, 17–32. [Google Scholar] [CrossRef]
- Lee, P.H.; Tu, C.T.; Hsiao, C.C.; Tsai, M.S.; Ho, C.M.; Cheng, N.C.; Hung, T.M.; Shih, D.T. Antifibrotic Activity of Human Placental Amnion Membrane-Derived CD34+ Mesenchymal Stem/Progenitor Cell Transplantation in Mice With Thioacetamide-Induced Liver Injury. Stem Cells Transl. Med. 2016, 5, 1473–1484. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.W.; Zhang, H.Z.; Guo, L.; Kim, J.M.; Kim, M.H. Amniotic mesenchymal stem cells enhance wound healing in diabetic NOD/SCID mice through high angiogenic and engraftment capabilities. PLoS ONE 2012, 7, e41105. [Google Scholar] [CrossRef]
- Tuca, A.C.; Ertl, J.; Hingerl, K.; Pichlsberger, M.; Fuchs, J.; Wurzer, P.; Pfeiffer, D.; Bubalo, V.; Parvizi, D.; Kamolz, L.P.; et al. Comparison of Matrigel and Matriderm as a carrier for human amnion-derived mesenchymal stem cells in wound healing. Placenta 2016, 48, 99–103. [Google Scholar] [CrossRef]
- Magatti, M.; Vertua, E.; De Munari, S.; Caro, M.; Caruso, M.; Silini, A.; Delgado, M.; Parolini, O. Human amnion favours tissue repair by inducing the M1-to-M2 switch and enhancing M2 macrophage features. J. Tissue Eng. Regen. Med. 2017, 11, 2895–2911. [Google Scholar] [CrossRef]
- Parolini, O.; Souza-Moreira, L.; O’Valle, F.; Magatti, M.; Hernandez-Cortes, P.; Gonzalez-Rey, E.; Delgado, M. Therapeutic effect of human amniotic membrane-derived cells on experimental arthritis and other inflammatory disorders. Arthritis Rheumatol. 2014, 66, 327–339. [Google Scholar] [CrossRef]
- Shu, J.; Pan, L.; Huang, X.; Wang, P.; Li, H.; He, X.; Cai, Z. Transplantation of human amnion mesenchymal cells attenuates the disease development in rats with collagen-induced arthritis. Clin. Exp. Rheumatol. 2015, 33, 484–490. [Google Scholar]
- Onishi, R.; Ohnishi, S.; Higashi, R.; Watari, M.; Yamahara, K.; Okubo, N.; Nakagawa, K.; Katsurada, T.; Suda, G.; Natsuizaka, M.; et al. Human Amnion-Derived Mesenchymal Stem Cell Transplantation Ameliorates Dextran Sulfate Sodium-Induced Severe Colitis in Rats. Cell Transplant. 2015, 24, 2601–2614. [Google Scholar] [CrossRef] [Green Version]
- Pischiutta, F.; Brunelli, L.; Romele, P.; Silini, A.; Sammali, E.; Paracchini, L.; Marchini, S.; Talamini, L.; Bigini, P.; Boncoraglio, G.B.; et al. Protection of Brain Injury by Amniotic Mesenchymal Stromal Cell-Secreted Metabolites. Crit. Care Med. 2016, 44, e1118–e1131. [Google Scholar] [CrossRef]
- Giampa, C.; Alvino, A.; Magatti, M.; Silini, A.R.; Cardinale, A.; Paldino, E.; Fusco, F.R.; Parolini, O. Conditioned medium from amniotic cells protects striatal degeneration and ameliorates motor deficits in the R6/2 mouse model of Huntington’s disease. J. Cell. Mol. Med. 2019, 23, 1581–1592. [Google Scholar] [CrossRef]
- Pianta, S.; Bonassi Signoroni, P.; Muradore, I.; Rodrigues, M.F.; Rossi, D.; Silini, A.; Parolini, O. Amniotic membrane mesenchymal cells-derived factors skew T cell polarization toward Treg and downregulate Th1 and Th17 cells subsets. Stem Cell Rev. 2015, 11, 394–407. [Google Scholar] [CrossRef] [Green Version]
- Pianta, S.; Magatti, M.; Vertua, E.; Bonassi Signoroni, P.; Muradore, I.; Nuzzo, A.M.; Rolfo, A.; Silini, A.; Quaglia, F.; Todros, T.; et al. Amniotic mesenchymal cells from pre-eclamptic placentae maintain immunomodulatory features as healthy controls. J. Cell. Mol. Med. 2016, 20, 157–169. [Google Scholar] [CrossRef]
- Magatti, M.; Caruso, M.; De Munari, S.; Vertua, E.; De, D.; Manuelpillai, U.; Parolini, O. Human Amniotic Membrane-Derived Mesenchymal and Epithelial Cells Exert Different Effects on Monocyte-Derived Dendritic Cell Differentiation and Function. Cell Transplant. 2015, 24, 1733–1752. [Google Scholar] [CrossRef]
- Li, J.; Koike-Soko, C.; Sugimoto, J.; Yoshida, T.; Okabe, M.; Nikaido, T. Human Amnion-Derived Stem Cells Have Immunosuppressive Properties on NK Cells and Monocytes. Cell Transplant. 2015, 24, 2065–2076. [Google Scholar] [CrossRef] [Green Version]
- Riboh, J.C.; Saltzman, B.M.; Yanke, A.B.; Cole, B.J. Human Amniotic Membrane-Derived Products in Sports Medicine: Basic Science, Early Results, and Potential Clinical Applications. Am. J. Sports Med. 2016, 44, 2425–2434. [Google Scholar] [CrossRef]
- Heckmann, N.; Auran, R.; Mirzayan, R. Application of Amniotic Tissue in Orthopedic Surgery. Am. J. Orthop. (Belle Mead NJ) 2016, 45, E421–E425. [Google Scholar]
- Nicodemo, M.C.; Neves, L.R.; Aguiar, J.C.; Brito, F.S.; Ferreira, I.; Sant’Anna, L.B.; Raniero, L.J.; Martins, R.Á.; Barja, P.R.; Arisawa, E.A. Amniotic membrane as an option for treatment of acute Achilles tendon injury in rats. Acta Cir. Bras. 2017, 32, 125–139. [Google Scholar] [CrossRef] [Green Version]
- Kueckelhaus, M.; Philip, J.; Kamel, R.A.; Canseco, J.A.; Hackl, F.; Kiwanuka, E.; Kim, M.J.; Wilkie, R.; Caterson, E.J.; Junker, J.P.; et al. Sustained release of amnion-derived cellular cytokine solution facilitates achilles tendon healing in rats. Eplasty 2014, 14, e29. [Google Scholar]
- Philip, J.; Hackl, F.; Canseco, J.A.; Kamel, R.A.; Kiwanuka, E.; Diaz-Siso, J.R.; Caterson, E.J.; Junker, J.P.; Eriksson, E. Amnion-derived multipotent progenitor cells improve achilles tendon repair in rats. Eplasty 2013, 13, e31. [Google Scholar]
- Lange-Consiglio, A.; Perrini, C.; Tasquier, R.; Deregibus, M.C.; Camussi, G.; Pascucci, L.; Marini, M.G.; Corradetti, B.; Bizzaro, D.; De Vita, B.; et al. Equine Amniotic Microvesicles and Their Anti-Inflammatory Potential in a Tenocyte Model In Vitro. Stem Cells Dev. 2016, 25, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Lange-Consiglio, A.; Lazzari, B.; Perrini, C.; Pizzi, F.; Stella, A.; Cremonesi, F.; Capra, E. MicroRNAs of Equine Amniotic Mesenchymal Cell-derived Microvesicles and Their Involvement in Anti-inflammatory Processes. Cell Transplant. 2018, 27, 45–54. [Google Scholar] [CrossRef] [Green Version]
- D’haene, B.; Mestdagh, P.; Hellemans, J.; Vandesompele, J. miRNA expression profiling: From reference genes to global mean normalization. Methods Mol. Biol. 2012, 822, 261–272. [Google Scholar] [CrossRef]
- Mestdagh, P.; Van Vlierberghe, P.; De Weer, A.; Muth, D.; Westermann, F.; Speleman, F.; Vandesompele, J. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009, 10, R64. [Google Scholar] [CrossRef] [Green Version]
- Ragni, E.; De Luca, P.; Perucca Orfei, C.; Colombini, A.; Viganò, M.; Lugano, G.; Bollati, V.; de Girolamo, L. Insights into Inflammatory Priming of Adipose-Derived Mesenchymal Stem Cells: Validation of Extracellular Vesicles-Embedded miRNA Reference Genes as A Crucial Step for Donor Selection. Cells 2019, 23, 369. [Google Scholar] [CrossRef] [Green Version]
- Ragni, E.; Perucca Orfei, C.; De Luca, P.; Colombini, A.; Viganò, M.; Lugano, G.; Bollati, V.; de Girolamo, L. Identification of miRNA Reference Genes in Extracellular Vesicles from Adipose Derived Mesenchymal Stem Cells for Studying Osteoarthritis. Int. J. Mol. Sci. 2019, 20, 1108. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, RESEARCH0034. [Google Scholar] [CrossRef] [Green Version]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Dubin, A.; Greenberg, D.R.; Iglinski-Benjamin, K.C.; Abrams, G.D. Effect of micro-RNA on tenocytes and tendon-related gene expression: A systematic review. J. Orthop. Res. 2018, 36, 2823–2829. [Google Scholar] [CrossRef]
- Dean, B.J.F.; Dakin, S.G.; Millar, N.L.; Carr, A.J. Review: Emerging concepts in the pathogenesis of tendinopathy. Surgeon 2017, 15, 349–354. [Google Scholar] [CrossRef]
- Steinmann, S.; Pfeifer, C.G.; Brochhausen, C.; Docheva, D. Spectrum of Tendon Pathologies: Triggers, Trails and End-State. Int. J. Mol. Sci. 2020, 21, 844. [Google Scholar] [CrossRef] [Green Version]
- Magatti, M.; Pianta, S.; Silini, A.; Parolini, O. Isolation, Culture, and Phenotypic Characterization of Mesenchymal Stromal Cells from the Amniotic Membrane of the Human Term Placenta. Methods Mol. Biol. 2016, 1416, 233–244. [Google Scholar] [CrossRef]
- Webber, J.; Clayton, A. How pure are your vesicles? J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Liu, F.; Xiang, G.; Jiang, D.; Pu, X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis. Markers 2015, 2015, 893594. [Google Scholar] [CrossRef]
- Li, Y.; Xiang, G.M.; Liu, L.L.; Liu, C.; Liu, F.; Jiang, D.N.; Pu, X.Y. Assessment of endogenous reference gene suitability for serum exosomal microRNA expression analysis in liver carcinoma resection studies. Mol. Med. Rep. 2015, 12, 4683–4691. [Google Scholar] [CrossRef] [Green Version]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [Green Version]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef] [Green Version]
- Lange, T.; Stracke, S.; Rettig, R.; Lendeckel, U.; Kuhn, J.; Schlüter, R.; Rippe, V.; Endlich, K.; Endlich, N. Identification of miR-16 as an endogenous reference gene for the normalization of urinary exosomal miRNA expression data from CKD patients. PLoS ONE 2017, 12, e0183435. [Google Scholar] [CrossRef]
- Gouin, K.; Peck, K.; Antes, T.; Johnson, J.L.; Li, C.; Vaturi, S.D.; Middleton, R.; de Couto, G.; Walravens, A.S.; Rodriguez-Borlado, L.; et al. A comprehensive method for identification of suitable reference genes in extracellular vesicles. J. Extracell. Vesicles 2017, 6, 1347019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santovito, D.; De Nardis, V.; Marcantonio, P.; Mandolini, C.; Paganelli, C.; Vitale, E.; Buttitta, F.; Bucci, M.; Mezzetti, A.; Consoli, A.; et al. Plasma exosome microRNA profiling unravels a new potential modulator of adiponectin pathway in diabetes: Effect of glycemic control. J. Clin. Endocrinol. Metab. 2014, 99, E1681–E1685. [Google Scholar] [CrossRef]
- Lv, C.; Yang, T. Effective enrichment of urinary exosomes by polyethylene glycol for RNA detection. Biomed. Res. 2018, 29. [Google Scholar] [CrossRef] [Green Version]
- Millar, N.L.; Watts, A.E.; Akbar, M.; Hughes, T.; Kitson, S.; Gilchrist, D.S. MicroRNA-29a in equine tendinopathy—A translational target. Equine Vet. J. 2016, 48, 27. [Google Scholar]
- Watts, A.E.; Millar, N.L.; Platt, J.; Kitson, S.M.; Akbar, M.; Rech, R.; Griffin, J.; Pool, R.; Hughes, T.; McInnes, I.B.; et al. MicroRNA29a Treatment Improves Early Tendon Injury. Mol. Ther. 2017, 25, 2415–2426. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Wang, G.D.; Liu, J.P.; Wang, H.S.; Liu, X.M.; Wang, Q.; Cai, X.H. miR-135a modulates tendon stem/progenitor cell senescence via suppressing ROCK1. Bone 2015, 71, 210–216. [Google Scholar] [CrossRef]
- Thankam, F.G.; Boosani, C.S.; Dilisio, M.F.; Agrawal, D.K. MicroRNAs associated with inflammation in shoulder tendinopathy and glenohumeral arthritis. Mol. Cell. Biochem. 2018, 437, 81–97. [Google Scholar] [CrossRef]
- Geng, Y.; Zhao, X.; Xu, J.; Zhang, X.; Hu, G.; Fu, S.C.; Dai, K.; Chen, X.; Patrick, Y.S.; Zhang, X. Overexpression of mechanical sensitive miR-337-3p alleviates ectopic ossification in rat tendinopathy model via targeting IRS1 and Nox4 of tendon derived stem cells. J. Mol. Cell Biol. 2019, mjz030. [Google Scholar] [CrossRef] [Green Version]
- Cavalleri, T.; Angelici, L.; Favero, C.; Dioni, L.; Mensi, C.; Bareggi, C.; Palleschi, A.; Rimessi, A.; Consonni, D.; Bordini, L.; et al. Plasmatic extracellular vesicle microRNAs in malignant pleural mesothelioma and asbestos-exposed subjects suggest a 2-miRNA signature as potential biomarker of disease. PLoS ONE 2017, 12, e0176680. [Google Scholar] [CrossRef] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Ramos, L.T.; Sánchez-Abarca, L.I.; Muntión, S.; Preciado, S.; Puig, N.; López-Ruano, G.; Hernández-Hernández, Á.; Redondo, A.; Ortega, R.; Rodríguez, C.; et al. MSC surface markers (CD44, CD73, and CD90) can identify human MSC-derived extracellular vesicles by conventional flow cytometry. Cell Commun. Signal. 2016, 14, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barilani, M.; Peli, V.; Cherubini, A.; Dossena, M.; Dolo, V.; Lazzari, L. NG2 as an Identity and Quality Marker of Mesenchymal Stem Cell Extracellular Vesicles. Cells 2019, 8, 1524. [Google Scholar] [CrossRef] [Green Version]
- Avendaño-Félix, M.; Fuentes-Mera, L.; Ramos-Payan, R.; Aguilar-Medina, M.; Pérez-Silos, V.; Moncada-Saucedo, N.; Marchat, L.A.; González-Barrios, J.A.; Ruiz-García, E.; Astudillo-de la Vega, H.; et al. A Novel OsteomiRs Expression Signature for Osteoblast Differentiation of Human Amniotic Membrane-Derived Mesenchymal Stem Cells. Biomed. Res. Int. 2019, 2019, 8987268. [Google Scholar] [CrossRef] [Green Version]
- Megraw, M.; Sethupathy, P.; Corda, B.; Hatzigeorgiou, A.G. miRGen: A database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007, 35, D149–D155. [Google Scholar] [CrossRef] [Green Version]
- Schwarzenbach, H.; da Silva, A.M.; Calin, G.; Pantel, K. Data Normalization Strategies for MicroRNA Quantification. Clin. Chem. 2015, 61, 1333–1342. [Google Scholar] [CrossRef]
- Meyer, S.U.; Pfaffl, M.W.; Ulbrich, S.E. Normalization strategies for microRNA profiling experiments: A ’normal’ way to a hidden layer of complexity? Biotechnol. Lett. 2010, 32, 1777–1788. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Gray, W.D.; French, K.M.; Ghosh-Choudhary, S.; Maxwell, J.T.; Brown, M.E.; Platt, M.O.; Searles, C.D.; Davis, M.E. Identification of therapeutic covariant microRNA clusters in hypoxia-treated cardiac progenitor cell exosomes using systems biology. Circ. Res. 2015, 116, 255–263. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Lombaert, I.M.; Hauser, B.R.; Patel, V.N.; Hoffman, M.P. Exosomal MicroRNA Transport from Salivary Mesenchyme Regulates Epithelial Progenitor Expansion during Organogenesis. Dev. Cell 2017, 40, 95–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, Q.; Zhang, J.; Li, C.; Miao, Y.R.; Lei, Q.; Li, Q.; Guo, A.Y. EVmiRNA: A database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019, 47, D89–D93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pathan, M.; Fonseka, P.; Chitti, S.V.; Kang, T.; Sanwlani, R.; Van Deun, J.; Hendrix, A.; Mathivanan, S. Vesiclepedia 2019: A compendium of RNA, proteins, lipids and metabolites in extracellular vesicles. Nucleic Acids Res. 2019, 47, D516–D519. [Google Scholar] [CrossRef] [Green Version]
- Valera, E.; Spencer, B.; Mott, J.; Trejo, M.; Adame, A.; Mante, M.; Rockenstein, E.; Troncoso, J.C.; Beach, T.G.; Masliah, E.; et al. MicroRNA-101 Modulates Autophagy and Oligodendroglial Alpha-Synuclein Accumulation in Multiple System Atrophy. Front. Mol. Neurosci. 2017, 10, 329. [Google Scholar] [CrossRef]
- Wang, L.; Zhuang, L.; Rong, H.; Guo, Y.; Ling, X.; Wang, R.; Yu, X.; Zhang, W. MicroRNA-101 inhibits proliferation of pulmonary microvascular endothelial cells in a rat model of hepatopulmonary syndrome by targeting the JAK2/STAT3 signaling pathway. Mol. Med. Rep. 2015, 12, 8261–8267. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Wang, K.; Hu, F.; Qian, C.; Guan, H.; Feng, K.; Zhou, Y.; Chen, Z. MicroRNA-101 protects cardiac fibroblasts from hypoxia-induced apoptosis via inhibition of the TGF-β signaling pathway. Int. J. Biochem. Cell Biol. 2015, 65, 155–164. [Google Scholar] [CrossRef]
- Xie, Y.; Yao, Q.; Butt, A.M.; Guo, J.; Tian, Z.; Bao, X.; Li, H.; Meng, Q.; Lu, J. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol. Ther. 2014, 15, 1248–1255. [Google Scholar] [CrossRef] [Green Version]
- Long, J.M.; Lahiri, D.K. MicroRNA-101 downregulates Alzheimer’s amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem. Biophys. Res. Commun. 2011, 404, 889–895. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Xiao, X.; Yang, Y.; Mishra, A.; Liang, Y.; Zeng, X.; Yang, X.; Xu, D.; Blackburn, M.R.; Henke, C.A.; et al. MicroRNA-101 attenuates pulmonary fibrosis by inhibiting fibroblast proliferation and activation. J. Biol. Chem. 2017, 292, 16420–16439. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Hua, R.; Gong, Z.; Shang, B.; Huang, Y.; Guo, L.; Liu, T.; Xue, J. Human amniotic epithelial cells inhibit CD4+ T cell activation in acute kidney injury patients by influencing the miR-101-c-Rel-IL-2 pathway. Mol. Immunol. 2017, 81, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Floris, I.; Descamps, B.; Vardeu, A.; Mitić, T.; Posadino, A.M.; Shantikumar, S.; Sala-Newby, G.; Capobianco, G.; Mangialardi, G.; Howard, L.; et al. Gestational diabetes mellitus impairs fetal endothelial cell functions through a mechanism involving microRNA-101 and histone methyltransferase enhancer of zester homolog-2. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 664–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.Z.; Deng, F.; Li, H.; Wang, D.D.; Zhang, W.; Ding, L.; Tang, J.H. MiR-101: A potential therapeutic target of cancers. Am. J. Transl. Res. 2018, 10, 3310–3321. [Google Scholar] [PubMed]
- Knyazev, E.N.; Samatov, T.R.; Fomicheva, K.A.; Nyushko, K.M.; Alekseev, B.Y.; Shkurnikov, M.Y. MicroRNA hsa-miR-4674 in Hemolysis-Free Blood Plasma Is Associated with Distant Metastases of Prostatic Cancer. Bull. Exp. Biol. Med. 2016, 161, 112–115. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Ning, S.; Li, Z.; Qin, X.; Xu, W. miR-22 is down-regulated in esophageal squamous cell carcinoma and inhibits cell migration and invasion. Cancer Cell Int. 2014, 14, 138. [Google Scholar] [CrossRef] [Green Version]
- Damavandi, Z.; Torkashvand, S.; Vasei, M.; Soltani, B.M.; Tavallaei, M.; Mowla, S.J. Aberrant Expression of Breast Development-Related MicroRNAs, miR-22, miR-132, and miR-212, in Breast Tumor Tissues. J. Breast Cancer 2016, 19, 148–155. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Cong, S. The Emerging Role of microRNAs in Polyglutamine Diseases. Front. Mol. Neurosci. 2019, 12, 156. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Ding, M.; Zhang, H.; Xu, X.; Tang, J. Molecular mechanisms and clinical applications of miR-22 in regulating malignant progression in human cancer (Review). Int. J. Oncol. 2017, 50, 345–355. [Google Scholar] [CrossRef] [PubMed]



| Accession Number | Gene Name | Target Sequence (5’–3’) | Reference |
|---|---|---|---|
| MIMAT0000062 | let-7a-5p | UGAGGUAGUAGGUUGUAUAGUU | [48,49,50] |
| MIMAT0000069 | miR-16-5p | UAGCAGCACGUAAAUAUUGGCG | [51,52] |
| MIMAT0004495 | miR-22-5p | AGUUCUUCAGUGGCAAGCUUUA | [8] |
| MIMAT0000078 | miR-23a-3p | AUCACAUUGCCAGGGAUUUCC | [53] |
| MIMAT0000082 | miR-26a-5p | UUCAAGUAAUCCAGGAUAGGCU | [49,53] |
| MIMAT0004503 | miR-29a-5p | ACUGAUUUCUUUUGGUGUUCAG | [8] |
| MIMAT0000099 | miR-101-3p | UACAGUACUGUGAUAACUGAA | [53] |
| MIMAT0000101 | miR-103a-3p | AGCAGCAUUGUACAGGGCUAUGA | [48] |
| MIMAT0000278 | miR-221-3p | AGCUACAUUGUCUGCUGGGUUUC | [48,49] |
| MIMAT0004748 | miR-423-5p | UGAGGGGCAGAGAGCGAGACUUU | [54] |
| MIMAT0003393 | miR-425-5p | AAUGACACGAUCACUCCCGUUGA | [54] |
| MIMAT0003338 | miR-660-5p | UACCCAUUGCAUAUCGGAGUUG | [8] |
| NR_004394.1 | U6 snRNA | GUGCUCGCUUCGGCAGCACAUAUACUAAAAU UGGAACGATACAGAGAAGAUUAGCAUGGCCC CUGCGCAAGGAUGACACGCAAAUUCGUGAAG CGUUCCAUAUUUU | [55] |
| miRNA targets | |||
| MIMAT0000086 | miR-29a-3p | UAGCACCAUCUGAAAUCGGUUA | [56,57] |
| MIMAT0000428 | miR-135a-5p | UAUGGCUUUUUAUUCCUAUGUGA | [58] |
| MIMAT0000449 | miR-146a-5p | UGAGAACUGAAUUCCAUGGGUU | [59] |
| MIMAT0000754 | miR-337-3p | CUCCUAUAUGAUGCCUUUCUUC | [60] |
| MIMAT0000065 | let-7d-5p | AGAGGUAGUAGGUUGCAUAGUU | [59] |
| Gene Name | Genorm M-Value | Normfinder SV | Bestkeeper SD | Delta Ct SD | Geomean | Ranking Order |
|---|---|---|---|---|---|---|
| miR-101-3p | 0.19 (1) | 0.09 (1) | 0.18 (2) | 0.68 (1) | 1.32 | 1 |
| miR-22-5p | 0.19 (1) | 0.10 (2) | 0.17 (1) | 0.69 (2) | 1.41 | 2 |
| miR-221-3p | 0.28 (3) | 0.24 (4) | 0.18 (3) | 0.74 (3) | 2.91 | 3 |
| miR-660-5p | 0.37 (4) | 0.16 (3) | 0.26 (4) | 0.76 (4) | 3.72 | 4 |
| miR-29a-5p | 0.46 (5) | 0.66 (5) | 0.40 (5) | 0.93 (5) | 5 | 5 |
| miR-16-5p | 0.59 (6) | 0.84 (6) | 0.65 (6) | 1.03 (6) | 6 | 6 |
| miR-26a-5p | 0.66 (7) | 1.02 (7) | 0.71 (7) | 1.13 (7) | 7 | 7 |
| miR-423-5p | 0.85 (8) | 1.17 (8) | 0.81 (8) | 1.28 (8) | 8 | 8 |
| U6 snRNA | 0.95 (9) | 1.18 (9) | 0.82 (9) | 1.29 (9) | 9 | 9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ragni, E.; Perucca Orfei, C.; Silini, A.R.; Colombini, A.; Viganò, M.; Parolini, O.; de Girolamo, L. miRNA Reference Genes in Extracellular Vesicles Released from Amniotic Membrane-Derived Mesenchymal Stromal Cells. Pharmaceutics 2020, 12, 347. https://doi.org/10.3390/pharmaceutics12040347
Ragni E, Perucca Orfei C, Silini AR, Colombini A, Viganò M, Parolini O, de Girolamo L. miRNA Reference Genes in Extracellular Vesicles Released from Amniotic Membrane-Derived Mesenchymal Stromal Cells. Pharmaceutics. 2020; 12(4):347. https://doi.org/10.3390/pharmaceutics12040347
Chicago/Turabian StyleRagni, Enrico, Carlotta Perucca Orfei, Antonietta Rosa Silini, Alessandra Colombini, Marco Viganò, Ornella Parolini, and Laura de Girolamo. 2020. "miRNA Reference Genes in Extracellular Vesicles Released from Amniotic Membrane-Derived Mesenchymal Stromal Cells" Pharmaceutics 12, no. 4: 347. https://doi.org/10.3390/pharmaceutics12040347