Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
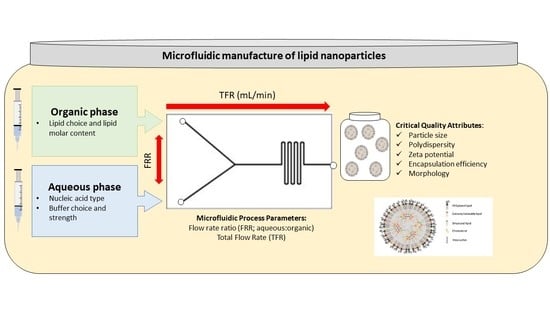
2.2. Microfluidics
2.3. Scalable Production of Lipid Nanoparticles Using a Toroidal Micromixer (TrM)
2.4. LNP Characterisation: Particle Size, Polydispersity and Zeta Potential
2.5. TNS Fluorescence Titration Assay
2.6. Quantification of Nucleic Acid Loading: Poly(A), ssDNA and mRNA
2.7. Cryogenic Transmission Electron Microscopy (Cryo-TEM)
2.8. Statistical Analysis
3. Results
3.1. Method Optimisation for Lipid Nanoparticles (LNPs)
3.1.1. Effect of Microfluidic Operating Parameters: Total Flow Rate and Flow Rate Ratio
3.1.2. Effect of the Lipid Molar Ratio
3.1.3. Effect of the Choice of Structural Lipid, Coating Lipid and Buffer on the Physicochemical Characteristics of DOTAP-cLNPs
3.2. Scalability of the Microfluidic Method for Production of iLNPs (GenVoy-ILM™)
3.3. Encapsulation of Nucleic Acids into LNPs
4. Discussions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Access
References
- Dammes, N.; Peer, D. Paving the road for RNA therapeutics. Trends Pharmacol. Sci. 2020, 41, 755–775. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Park, J.; Pei, Y.; Xu, J.; Yeo, Y. Pharmacokinetics and biodistribution of recently-developed siRNA nanomedicines. Adv. Drug Deliv. Rev. 2016, 104, 93–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaczmarek, J.C.; Kowalski, P.S.; Anderson, D.G. Advances in the delivery of RNA therapeutics: From concept to clinical reality. Genome Med. 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- U.S. Food and Drug Administration. Drug Approval Package: Onpattro (Patisiran). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2018/210922Orig1s000TOC.cfm (accessed on 29 September 2020).
- U.S. Food and Drug Administration. Drug Approval Package: GIVLAARI (Givosiran) Injection. Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2019/212194Orig1s000TOC.cfm (accessed on 29 September 2020).
- Semple, S.C.; Klimuk, S.K.; Harasym, T.O.; Dos Santos, N.; Ansell, S.M.; Wong, K.F.; Maurer, N.; Stark, H.; Cullis, P.R.; Hope, M.J.; et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: Formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta (BBA) Biomembr. 2001, 1510, 152–166. [Google Scholar] [CrossRef] [Green Version]
- Jayaraman, M.; Ansell, S.M.; Mui, B.L.; Tam, Y.K.; Chen, J.; Du, X.; Butler, D.; Eltepu, L.; Matsuda, S.; Narayanannair, J.K.; et al. Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. Int. Ed. 2012, 51, 8529–8533. [Google Scholar] [CrossRef]
- Akinc, A.; Maier, M.A.; Manoharan, M.; Fitzgerald, K.; Jayaraman, M.; Barros, S.; Ansell, S.; Du, X.; Hope, M.J.; Madden, T.D. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019, 14, 1084–1087. [Google Scholar] [CrossRef]
- Samaridou, E.; Heyes, J.; Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020. [Google Scholar] [CrossRef]
- MacLachlan, I. Liposomal formulations for nucleic acid delivery. Antisense Drug Technol. Princ. Strat. Appl. 2007, 2, 237–270. [Google Scholar]
- Jahn, A.; Vreeland, W.N.; Gaitan, M.; Locascio, L.E. Controlled Vesicle Self-Assembly in Microfluidic Channels with Hydrodynamic Focusing. J. Am. Chem. Soc. 2004, 126, 2674–2675. [Google Scholar] [CrossRef]
- Wagner, A.; Vorauer-Uhl, K.; Kreismayr, G.; Katinger, H. The crossflow injection technique: An improvement of the ethanol injection method. J. Liposome Res. 2002, 12, 259–270. [Google Scholar] [CrossRef]
- Lou, G.; Anderluzzi, G.; Schmidt, S.T.; Woods, S.; Gallorini, S.; Brazzoli, M.; Giusti, F.; Ferlenghi, I.; Johnson, R.N.; Roberts, C.W.; et al. Delivery of self-amplifying mRNA vaccines by cationic lipid nanoparticles: The impact of cationic lipid selection. J. Controlled Rel. 2020, 325, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.; Forbes, N.; Roces, C.B.; Anderluzzi, G.; Lou, G.; Abraham, S.; Ingalls, L.; Marshall, K.; Leaver, T.J.; Watts, J.A.; et al. Using microfluidics for scalable manufacturing of nanomedicines from bench to GMP: A case study using protein-loaded liposomes. Int. J. Pharm. 2020, 582, 119266. [Google Scholar] [CrossRef] [PubMed]
- Geall, A.J.; Verma, A.; Otten, G.R.; Shaw, C.A.; Hekele, A.; Banerjee, K.; Cu, Y.; Beard, C.W.; Brito, L.A.; Krucker, T.; et al. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA 2012, 109, 14604–14609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belliveau, N.M.; Huft, J.; Lin, P.J.C.; Chen, S.; Leung, A.K.K.; Leaver, T.J.; Wild, A.W.; Lee, J.B.; Taylor, R.J.; Tam, Y.K.; et al. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol. Ther. Nucleic Acids 2012, 1, e37. [Google Scholar] [CrossRef]
- Hassett, K.J.; Benenato, K.E.; Jacquinet, E.; Lee, A.; Woods, A.; Yuzhakov, O.; Himansu, S.; Deterling, J.; Geilich, B.M.; Ketova, T.; et al. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids 2019, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kauffman, K.J.; Dorkin, J.R.; Yang, J.H.; Heartlein, M.W.; DeRosa, F.; Mir, F.F.; Fenton, O.S.; Anderson, D.G. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015, 15, 7300–7306. [Google Scholar] [CrossRef]
- Leung, A.K.; Tam, Y.Y.; Chen, S.; Hafez, I.M.; Cullis, P.R. Microfluidic Mixing: A General Method for Encapsulating Macromolecules in Lipid Nanoparticle Systems. J. Phys. Chem. B 2015, 119, 8698–8706. [Google Scholar] [CrossRef]
- Maeki, M.; Saito, T.; Sato, Y.; Yasui, T.; Kaji, N.; Ishida, A.; Tani, H.; Baba, Y.; Harashima, H.; Tokeshi, M. A strategy for synthesis of lipid nanoparticles using microfluidic devices with a mixer structure. RSC Adv. 2015, 5, 46181–46185. [Google Scholar] [CrossRef] [Green Version]
- Roces, C.B.; Khadke, S.; Christensen, D.; Perrie, Y. Scale-Independent Microfluidic Production of Cationic Liposomal Adjuvants and Development of Enhanced Lymphatic Targeting Strategies. Mol. Pharm. 2019. [Google Scholar] [CrossRef] [Green Version]
- Forbes, N.; Hussain, M.T.; Briuglia, M.L.; Edwards, D.P.; Horst, J.H.t.; Szita, N.; Perrie, Y. Rapid and scale-independent microfluidic manufacture of liposomes entrapping protein incorporating in-line purification and at-line size monitoring. Int. J. Pharm. 2019, 556, 68–81. [Google Scholar] [CrossRef] [Green Version]
- Roces, C.B.; Port, E.C.; Daskalakis, N.N.; Watts, J.A.; Aylott, J.W.; Halbert, G.W.; Perrie, Y. Rapid scale-up and production of active-loaded PEGylated liposomes. Int. J. Pharm. 2020, 586, 119566. [Google Scholar] [CrossRef] [PubMed]
- Roces, C.B.; Christensen, D.; Perrie, Y. Translating the fabrication of protein-loaded poly (lactic-co-glycolic acid) nanoparticles from bench to scale-independent production using microfluidics. Drug Deliv. Transl. Res. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Roces, C.B.; Hussain, M.T.; Schmidt, S.T.; Christensen, D.; Perrie, Y. Investigating Prime-Pull Vaccination through a Combination of Parenteral Vaccination and Intranasal Boosting. Vaccines 2019, 8, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderluzzi, G.; Lou, G.; Gallorini, S.; Brazzoli, M.; Johnson, R.; O’Hagan, D.T.; Baudner, B.C.; Perrie, Y. Investigating the Impact of Delivery System Design on the Efficacy of Self-Amplifying RNA Vaccines. Vaccines 2020, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Zizzari, A.; Bianco, M.; Carbone, L.; Perrone, E.; Amato, F.; Maruccio, G.; Rendina, F.; Arima, V. Continuous-flow production of injectable liposomes via a microfluidic approach. Materials 2017, 10, 1411. [Google Scholar] [CrossRef] [Green Version]
- Jahn, A.; Stavis, S.M.; Hong, J.S.; Vreeland, W.N.; DeVoe, D.L.; Gaitan, M. Microfluidic mixing and the formation of nanoscale lipid vesicles. ACS Nano 2010, 4, 2077–2087. [Google Scholar] [CrossRef]
- Jeffs, L.B.; Palmer, L.R.; Ambegia, E.G.; Giesbrecht, C.; Ewanick, S.; MacLachlan, I. A Scalable, Extrusion-Free Method for Efficient Liposomal Encapsulation of Plasmid DNA. Pharm. Res. 2005, 22, 362–372. [Google Scholar] [CrossRef] [Green Version]
- Kulkarni, J.A.; Myhre, J.L.; Chen, S.; Tam, Y.Y.C.; Danescu, A.; Richman, J.M.; Cullis, P.R. Design of lipid nanoparticles for in vitro and in vivo delivery of plasmid DNA. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1377–1387. [Google Scholar] [CrossRef]
- Cheng, X.; Lee, R.J. The role of helper lipids in lipid nanoparticles (LNPs) designed for oligonucleotide delivery. Adv. Drug Deliv. Rev. 2016, 99, 129–137. [Google Scholar] [CrossRef]
- Rodrigueza, W.V.; Wheeler, J.J.; Klimuk, S.K.; Kitson, C.N.; Hope, M.J. Transbilayer Movement and Net Flux of Cholesterol and Cholesterol Sulfate between Liposomal Membranes. Biochemistry 1995, 34, 6208–6217. [Google Scholar] [CrossRef]
- Sato, Y.; Okabe, N.; Note, Y.; Hashiba, K.; Maeki, M.; Tokeshi, M.; Harashima, H. Hydrophobic scaffolds of pH-sensitive cationic lipids contribute to miscibility with phospholipids and improve the efficiency of delivering short interfering RNA by small-sized lipid nanoparticles. Acta Biomater. 2020, 102, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Holland, J.W.; Hui, C.; Cullis, P.R.; Madden, T.D. Poly(ethylene glycol)−Lipid Conjugates Regulate the Calcium-Induced Fusion of Liposomes Composed of Phosphatidylethanolamine and Phosphatidylserine. Biochemistry 1996, 35, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, H.; Li, Y.; Leng, X.; Huang, W.; Ma, Y.; Xu, T.; Qi, X. Transfection reagent Lipofectamine triggers type I interferon signaling activation in macrophages. Immunol. Cell Biol. 2019, 97, 92–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Judge, A.; McClintock, K.; Phelps, J.R.; MacLachlan, I. Hypersensitivity and Loss of Disease Site Targeting Caused by Antibody Responses to PEGylated Liposomes. Mol. Ther. 2006, 13, 328–337. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Tam, Y.Y.C.; Lin, P.J.C.; Sung, M.M.H.; Tam, Y.K.; Cullis, P.R. Influence of particle size on the in vivo potency of lipid nanoparticle formulations of siRNA. J. Controlled Rel. 2016, 235, 236–244. [Google Scholar] [CrossRef]
- Mui, B.L.; Tam, Y.K.; Jayaraman, M.; Ansell, S.M.; Du, X.; Tam, Y.Y.C.; Lin, P.J.C.; Chen, S.; Narayanannair, J.K.; Rajeev, K.G.; et al. Influence of Polyethylene Glycol Lipid Desorption Rates on Pharmacokinetics and Pharmacodynamics of siRNA Lipid Nanoparticles. Mol. Ther. Nucleic Acids 2013, 2, e139. [Google Scholar] [CrossRef]
- Lou, G.; Anderluzzi, G.; Woods, S.; Roberts, C.W.; Perrie, Y. A novel microfluidic-based approach to formulate size-tuneable large unilamellar cationic liposomes: Formulation, cellular uptake and biodistribution investigations. Eur. J. Pharm. Biopharm. 2019, 143, 51–60. [Google Scholar] [CrossRef]
- Sato, Y.; Hatakeyama, H.; Hyodo, M.; Harashima, H. Relationship Between the Physicochemical Properties of Lipid Nanoparticles and the Quality of siRNA Delivery to Liver Cells. Mol. Ther. 2016, 24, 788–795. [Google Scholar] [CrossRef] [Green Version]
- Blakney, A.K.; McKay, P.F.; Yus, B.I.; Aldon, Y.; Shattock, R.J. Inside out: Optimization of lipid nanoparticle formulations for exterior complexation and in vivo delivery of saRNA. Gene Ther. 2019, 26, 363–372. [Google Scholar] [CrossRef]
- Kulkarni, J.A.; Witzigmann, D.; Leung, J.; Tam, Y.Y.C.; Cullis, P.R. On the role of helper lipids in lipid nanoparticle formulations of siRNA. Nanoscale 2019, 11, 21733–21739. [Google Scholar] [CrossRef]
- Leung, A.K.K.; Hafez, I.M.; Baoukina, S.; Belliveau, N.M.; Zhigaltsev, I.V.; Afshinmanesh, E.; Tieleman, D.P.; Hansen, C.L.; Hope, M.J.; Cullis, P.R. Lipid Nanoparticles Containing siRNA Synthesized by Microfluidic Mixing Exhibit an Electron-Dense Nanostructured Core. J. Phys. Chem. C 2012, 116, 18440–18450. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-Y.; Wang, W.-T.; Liu, C.-C.; Fu, L.-M. Passive mixers in microfluidic systems: A review. Chem. Eng. J. 2016, 288, 146–160. [Google Scholar] [CrossRef]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roces, C.B.; Lou, G.; Jain, N.; Abraham, S.; Thomas, A.; Halbert, G.W.; Perrie, Y. Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics. Pharmaceutics 2020, 12, 1095. https://doi.org/10.3390/pharmaceutics12111095
Roces CB, Lou G, Jain N, Abraham S, Thomas A, Halbert GW, Perrie Y. Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics. Pharmaceutics. 2020; 12(11):1095. https://doi.org/10.3390/pharmaceutics12111095
Chicago/Turabian StyleRoces, Carla B., Gustavo Lou, Nikita Jain, Suraj Abraham, Anitha Thomas, Gavin W. Halbert, and Yvonne Perrie. 2020. "Manufacturing Considerations for the Development of Lipid Nanoparticles Using Microfluidics" Pharmaceutics 12, no. 11: 1095. https://doi.org/10.3390/pharmaceutics12111095