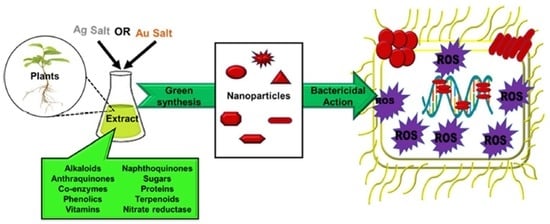
Bactericidal Antibacterial Mechanism of Plant Synthesized Silver, Gold and Bimetallic Nanoparticles
Abstract
:1. Introduction
2. Biogenic Synthesis
2.1. Silver Nanoparticles
2.2. Gold Nanoparticles
2.3. Ag-Au Bimetallic Nanoparticles
| S/N | Sample | Biogenic source/Extraction Method | Bioactive Compound | NPs | Size (nm) | Shape | Ref. |
|---|---|---|---|---|---|---|---|
| 1 | Coffea arabica | Seed/ Ethanolic Extraction at 60 °C for 1 h | Phenolics | Ag | 20–30 nm | Spheres and ellipsoidal | [34] |
| 2 | Olive tree | Leaf/Aqueous extraction by boiling for 10 min | Oleuropein | Ag | 20–25 nm | Spheres | [35] |
| 3 | Ocimum sanctum and quercetin | Leaf/Aqueous extraction at 60 °C for 10 min | Quercetin | Ag | 14.6 nm and 11.35 nm | Spheres | [36] |
| 4 | Origanum vulgare | Leaf/Aqueous extraction by reflux by for 4 h | Alkaloids, flavonoids, terpenoids | Ag | 2–25 | Spherical | [38] |
| 5 | Tagetes erecta (Marigold) | Flower/Aqueous extraction for 10 min | Flavonoids, saponins | Ag | 46.11 nm | Spheres | [39] |
| 6 | Combretum erythropyllum | Leaf/Aqueous extraction at 90 °C for 1 h | Flavonoids | Ag | 5–26 nm | Spherical | [18] |
| 7 | Mentha aquatica | Leaf/Aqueous extraction by sonication | Polyphenols, flavonoids | 8 nm | Spheres | [40] | |
| 8 | Punica granatum | Leaf/Ethanolic extraction for 48 h at room temperature | Polyphenols, flavonoids | Ag | 20–40 nm | Polygonal | [41] |
| 9 | Carica papaya and Catharanthus roseus | Leaf/Aqueous extraction at room temperature | Papain, α-tocopherol, alkaloids, flavonoids | Au | 6–18 nm | Spherical, Triangle, hexagonal | [43] |
| 10. | Citrullus lanatus rind (Watermelon) | Fruit/Aqueous extraction for 10 min by boiling | Citrulline, proteins, carotenoids | Au | 20–140 nm | Spheres | [44] |
| 11 | Cannabis sativa (Indian Hemp) Cortex and Xylem | Stem /Aqueous extraction for 10 min by boiling | Cannabinoids, terpenes, phenolics | Au | 12–18 nm and 20–40 nm | Spheres, rod, Triangle, hexagonal | [45] |
| 12. | Amomum villosum (Cardamom) | Fruit/Aqueous at 100 °C for 1 h via autoclave | *** | Au | 5–10 nm | Spheres | [46] |
| 13 | Pistacia atlantica | (Leaf and fruit)/Aqueous by boiling for 30 min | *** | Au | 50–60 | Spheres | [47] |
| 14 | Thyme | Leaf/Aqueous by boiling for 30 min | *** | Au | 6–26 nm | [48] | |
| 15 | Olea europaea fruit extract and Acacia nilotica husk | Fruit and husk/Aqueous extraction at room temperature | *** | Au | 44.96 nm | Spheres | [49] |
| 16 | Croton caudatus Geisel | Leaf/Aqueous extraction at 50 °C for 10 min | *** | Au | 20–50 nm | sphere | [50] |
| 17 | Beta vulgaris (Sugar beet) | Pulp/Aqueous purification | *** | Au | 50 nm | Nanowires | [51] |
| 18 | Garcina kola | Pulp/Aqueous extraction by boiling for 40 min | *** | Au | 18–38 nm | Spheres | [52] |
| 19 | Cryptolepis buchanani | Tea/Aqueous extraction at 60 °C for 15 min | Flavonoids, alkaloids, saponins, tannins | Au | 11.1 nm | Spheres | [53] |
20 | Solidago canadensis | Leaf/Aqueous extraction at 80 °C | Flavonoids, quercetin, saponins | Ag-Au | 15 nm | Spheres | [57] |
| 21 | Stigmaphyllon ovatum | Leaf | *** | Ag-Au | 14.9 nm | Spheres | [59] |
| 22 | Gloriosa superba | leaf/Aqueous extraction at 60 °C for 50 min | Superbine, colchicine, phytosterils, stigmasterin | Ag-Au | 10–20 nm | Spheres | [60] |
| 23 | Pomegranate | Seed/Aqueous extraction | Phenolics | Ag-Au | 12 nm | Spheres Rods Pentagonal | [61] |
| 24 | Arabic gum | Stems and branches of Arabic Senegal tree/Aqueous dissolution | Arabinose, rhamnose, glucoronic acid, arabinogalact-an–protein complex | Ag-Au | 3.1 nm | Spheres | [62] |
| 25 | Moringa oleifera | Leaves/Aqueous extraction at 80 °C for 15 min | Niazimicin, 4-(α-L-rhamnosyloxy) benzyl isothiocyanate, β-sitosterol-3-O-β-D-glucopyranoside | Ag-Au | 11–25 nm | Spheres Triangles Hexagonal | [63] |
3. Bacterial Resistance and Mutations
4. Overview of the Bactericidal Mechanism
4.1. Cell Membrane: Lipid and Protein Interaction
4.2. Free Radical Generation
4.3. DNA Damage
5. Conclusions and Future Perspectives
Funding
Conflicts of Interest
References
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346–2353. [Google Scholar] [CrossRef] [Green Version]
- WHO. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 16 September 2020).
- WHO. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Shrivastava, S.R.; Shrivastava, P.S.; Ramasamy, J. World health organization releases global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. J. Med. Soc. 2018, 32, 76. [Google Scholar] [CrossRef]
- Guo, Z.; Chen, Y.; Wang, Y.; Jiang, H.; Wang, X. Advances and challenges in metallic nanomaterial synthesis and antibacterial applications. J. Mater. Chem. B 2020, 8, 4764–4777. [Google Scholar] [CrossRef] [PubMed]
- Årdal, C.; Balasegaram, M.; Laxminarayan, R.; McAdams, D.; Outterson, K.; Rex, J.H.; Sumpradit, N. Antibiotic development—Economic, regulatory and societal challenges. Nat. Rev. Microbiol. 2020, 18, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Pelgrift, R.Y.; Friedman, A.J. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv. Drug Deliv. Rev. 2013, 65, 1803–1815. [Google Scholar] [CrossRef]
- Holm, V.R.A.; Greve, M.M.; Holst, B. A theoretical investigation of the optical properties of metal nanoparticles in water for photo thermal conversion enhancement. Energy Convers. Manag. 2017, 149, 536–542. [Google Scholar] [CrossRef]
- Srinoi, P.; Chen, Y.T.; Vittur, V.; Marquez, M.D.; Lee, T.R. Bimetallic nanoparticles: Enhanced magnetic and optical properties for emerging biological applications. Appl. Sci. 2018, 8, 1106. [Google Scholar] [CrossRef] [Green Version]
- Yuan, P.; Ding, X.; Yang, Y.Y.; Xu, Q.H. Metal Nanoparticles for Diagnosis and Therapy of Bacterial Infection. Adv. Healthc. Mater. 2018, 7, 1701392. [Google Scholar] [CrossRef]
- Khan, S.A. Metal nanoparticles toxicity: Role of physicochemical aspects. In Metal Nanoparticles for Drug Delivery and Diagnostic Applications; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Torres-Sangiao, E.; Holban, A.M.; Gestal, M.C. Advanced nanobiomaterials: Vaccines, diagnosis and treatment of infectious diseases. Molecules 2016, 21, 867. [Google Scholar] [CrossRef] [Green Version]
- Rai, M.; Ingle, A.P.; Birla, S.; Yadav, A.; Santos, C.A. Dos Strategic role of selected noble metal nanoparticles in medicine. Crit. Rev. Microbiol. 2016, 42, 696–719. [Google Scholar]
- Tang, S.; Zheng, J. Antibacterial Activity of Silver Nanoparticles: Structural Effects. Adv. Healthc. Mater. 2018, 7, 1701503. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Wen, J.; Xiong, X.; Hu, Y. Shape effect on the antibacterial activity of silver nanoparticles synthesized via a microwave-assisted method. Environ. Sci. Pollut. Res. 2016, 23, 4489–4497. [Google Scholar] [CrossRef] [PubMed]
- Amendola, V.; Bakr, O.M.; Stellacci, F. A study of the surface plasmon resonance of silver nanoparticles by the discrete dipole approximation method: Effect of shape, size, structure, and assembly. Plasmonics 2010, 5, 85–97. [Google Scholar] [CrossRef]
- Wang, J.; Xie, Y.; Wang, L.; Tang, J.; Li, J.; Kocaefe, D.; Kocaefe, Y.; Zhang, Z.; Li, Y.; Chen, C. In vivo pharmacokinetic features and biodistribution of star and rod shaped gold nanoparticles by multispectral optoacoustic tomography. RSC Adv. 2015, 5, 7529–7538. [Google Scholar] [CrossRef]
- Jemilugba, O.T.; Sakho, E.H.M.; Parani, S.; Mavumengwana, V.; Oluwafemi, O.S. Green synthesis of silver nanoparticles using Combretum erythrophyllum leaves and its antibacterial activities. Colloid Interface Sci. Commun. 2019, 31, 100191. [Google Scholar] [CrossRef]
- Zhou, X.; Xu, C.; Jin, Y.; Li, B. Visual chiral recognition of D/L-leucine using cube-shaped gold nanoparticles as colorimetric probes. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 223, 117263. [Google Scholar] [CrossRef]
- Slavin, Y.N.; Asnis, J.; Häfeli, U.O.; Bach, H. Metal nanoparticles: Understanding the mechanisms behind antibacterial activity. J. Nanobiotechnology 2017, 15, 65. [Google Scholar] [CrossRef]
- Ahmad, S.; Munir, S.; Zeb, N.; Ullah, A.; Khan, B.; Ali, J.; Bilal, M.; Omer, M.; Alamzeb, M.; Salman, S.M.; et al. Green nanotechnology: A review on green synthesis of silver nanoparticles—An ecofriendly approach. Int. J. Nanomed. 2019, 14, 5087. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.F.; Liu, Z.G.; Shen, W.; Gurunathan, S. Silver nanoparticles: Synthesis, characterization, properties, applications, and therapeutic approaches. Int. J. Mol. Sci. 2016, 17, 1534. [Google Scholar] [CrossRef]
- Das, C.G.A.; Kumar, V.G.; Dhas, T.S.; Karthick, V.; Govindaraju, K.; Joselin, J.M.; Baalamurugan, J. Antibacterial activity of silver nanoparticles (biosynthesis): A short review on recent advances. Biocatal. Agric. Biotechnol. 2020, 27, 101593. [Google Scholar] [CrossRef]
- Mochochoko, T.; Oluwafemi, O.S.; Jumbam, D.N.; Songca, S.P. Green synthesis of silver nanoparticles using cellulose extracted from an aquatic weed; Water hyacinth. Carbohydr. Polym. 2013, 98, 290–294. [Google Scholar] [CrossRef]
- Mittal, A.K.; Chisti, Y.; Banerjee, U.C. Synthesis of metallic nanoparticles using plant extracts. Biotechnol. Adv. 2013, 31, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, A.D.; Gopal, K. Biosynthesis of silver and gold nanoparticles using Chenopodium album leaf extract. Colloids Surf. A Physicochem. Eng. Asp. 2010, 369, 27–33. [Google Scholar] [CrossRef]
- Roy, A.; Bulut, O.; Some, S.; Mandal, A.K.; Yilmaz, M.D. Green synthesis of silver nanoparticles: Biomolecule-nanoparticle organizations targeting antimicrobial activity. RSC Adv. 2019, 9, 2673–2702. [Google Scholar] [CrossRef] [Green Version]
- Rai, A.; Prabhune, A.; Perry, C.C. Antibiotic mediated synthesis of gold nanoparticles with potent antimicrobial activity and their application in antimicrobial coatings. J. Mater. Chem. 2010, 20, 6789–6798. [Google Scholar] [CrossRef] [Green Version]
- Rajeshkumar, S.; Malarkodi, C.; Vanaja, M.; Annadurai, G. Anticancer and enhanced antimicrobial activity of biosynthesizd silver nanoparticles against clinical pathogens. J. Mol. Struct. 2016, 1116, 165–173. [Google Scholar] [CrossRef]
- Veerapandian, M.; Yun, K. Functionalization of biomolecules on nanoparticles: Specialized for antibacterial applications. Appl. Microbiol. Biotechnol. 2011, 90, 1655–1667. [Google Scholar] [CrossRef]
- Akhtar, M.S.; Panwar, J.; Yun, Y.S. Biogenic synthesis of metallic nanoparticles by plant extracts. ACS Sustain. Chem. Eng. 2013, 1, 591–602. [Google Scholar] [CrossRef]
- Shanmuganathan, R.; Karuppusamy, I.; Saravanan, M.; Muthukumar, H.; Ponnuchamy, K.; Ramkumar, V.S.; Pugazhendhi, A. Synthesis of Silver Nanoparticles and their Biomedical Applications—A Comprehensive Review. Curr. Pharm. Des. 2019, 25, 2650–2660. [Google Scholar] [CrossRef]
- Ovais, M.; Ahmad, I.; Khalil, A.T.; Mukherjee, S.; Javed, R.; Ayaz, M.; Raza, A.; Shinwari, Z.K. Wound healing applications of biogenic colloidal silver and gold nanoparticles: Recent trends and future prospects. Appl. Microbiol. Biotechnol. 2018, 102, 4305–4318. [Google Scholar] [CrossRef]
- Dhand, V.; Soumya, L.; Bharadwaj, S.; Chakra, S.; Bhatt, D.; Sreedhar, B. Green synthesis of silver nanoparticles using Coffea arabica seed extract and its antibacterial activity. Mater. Sci. Eng. C 2016, 58, 36–43. [Google Scholar] [CrossRef]
- Khalil, M.M.H.; Ismail, E.H.; El-Baghdady, K.Z.; Mohamed, D. Green synthesis of silver nanoparticles using olive leaf extract and its antibacterial activity. Arab. J. Chem. 2014, 7, 1131–1139. [Google Scholar] [CrossRef] [Green Version]
- Jain, S.; Mehata, M.S. Medicinal Plant Leaf Extract and Pure Flavonoid Mediated Green Synthesis of Silver Nanoparticles and their Enhanced Antibacterial Property. Sci. Rep. 2017, 7, 15867. [Google Scholar] [CrossRef]
- Leela, A.; Vivekanandan, M. Tapping the unexploited plant resources for the synthesis of silver nanoparticles. Afr. J. Biotechnol. 2008, 7, 3162–31645. [Google Scholar] [CrossRef]
- Shaik, M.R.; Khan, M.; Kuniyil, M.; Al-Warthan, A.; Alkhathlan, H.Z.; Siddiqui, M.R.H.; Shaik, J.P.; Ahamed, A.; Mahmood, A.; Khan, M.; et al. Plant-Extract-Assisted green synthesis of silver nanoparticles using Origanum vulgare L. Extract and their microbicidal activities. Sustainability 2018, 10, 913. [Google Scholar] [CrossRef] [Green Version]
- Padalia, H.; Moteriya, P.; Chanda, S. Green synthesis of silver nanoparticles from marigold flower and its synergistic antimicrobial potential. Arab. J. Chem. 2015, 8, 732–741. [Google Scholar] [CrossRef] [Green Version]
- Nouri, A.; Tavakkoli Yaraki, M.; Lajevardi, A.; Rezaei, Z.; Ghorbanpour, M.; Tanzifi, M. Ultrasonic-assisted green synthesis of silver nanoparticles using Mentha aquatica leaf extract for enhanced antibacterial properties and catalytic activity. Colloids Interface Sci. Commun. 2020, 35, 100252. [Google Scholar] [CrossRef]
- Swilam, N.; Nematallah, K.A. Polyphenols profile of pomegranate leaves and their role in green synthesis of silver nanoparticles. Sci. Rep. 2020, 10, 14851. [Google Scholar] [CrossRef]
- Aromal, S.A.; Philip, D. Green synthesis of gold nanoparticles using Trigonella foenum-graecum and its size-dependent catalytic activity. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 97, 1–5. [Google Scholar] [CrossRef]
- Muthukumar, T.; Sambandam, B.; Aravinthan, A.; Sastry, T.P.; Kim, J.H. Green synthesis of gold nanoparticles and their enhanced synergistic antitumor activity using HepG2 and MCF7 cells and its antibacterial effects. Process Biochem. 2016, 51, 384–391. [Google Scholar] [CrossRef]
- Patra, J.K.; Baek, K.H. Novel green synthesis of gold nanoparticles using Citrullus lanatus rind and investigation of proteasome inhibitory activity, antibacterial, and antioxidant potential. Int. J. Nanomed. 2015, 10, 7253–7264. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Pandit, S.; Garnæs, J.; Tunjic, S.; Mokkapati, V.R.S.S.; Sultan, A.; Thygesen, A.; Mackevica, A.; Mateiu, R.V.; Daugaard, A.E.; et al. Green synthesis of gold and silver nanoparticles from Cannabis sativa (Industrial hemp) and their capacity for biofilm inhibition. Int. J. Nanomed. 2018, 13, 3571–3591. [Google Scholar] [CrossRef] [PubMed]
- Soshnikova, V.; Kim, Y.J.; Singh, P.; Huo, Y.; Markus, J.; Ahn, S.; Castro-Aceituno, V.; Kang, J.; Chokkalingam, M.; Mathiyalagan, R.; et al. Cardamom fruits as a green resource for facile synthesis of gold and silver nanoparticles and their biological applications. Artif. Cells Nanomed. Biotechnol. 2018, 46, 108–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamelian, M.; Hemmati, S.; Varmira, K.; Veisi, H. Green synthesis, antibacterial, antioxidant and cytotoxic effect of gold nanoparticles using Pistacia Atlantica extract. J. Taiwan Inst. Chem. Eng. 2018, 93, 21–30. [Google Scholar] [CrossRef]
- Hamelian, M.; Varmira, K.; Veisi, H. Green synthesis and characterizations of gold nanoparticles using Thyme and survey cytotoxic effect, antibacterial and antioxidant potential. J. Photochem. Photobiol. B Biol. 2018, 184, 71–79. [Google Scholar] [CrossRef]
- Awad, M.A.; Eisa, N.E.; Virk, P.; Hendi, A.A.; Ortashi, K.M.O.O.; Mahgoub, A.A.S.A.; Elobeid, M.A.; Eissa, F.Z. Green synthesis of gold nanoparticles: Preparation, characterization, cytotoxicity, and anti-bacterial activities. Mater. Lett. 2019, 256, 126608. [Google Scholar] [CrossRef]
- Kumar, P.V.; Kala, S.M.J.; Prakash, K.S. Green synthesis of gold nanoparticles using Croton Caudatus Geisel leaf extract and their biological studies. Mater. Lett. 2019, 236, 19–22. [Google Scholar] [CrossRef]
- Castro, L.; Blázquez, M.L.; Muñoz, J.A.; González, F.; García-Balboa, C.; Ballester, A. Biosynthesis of gold nanowires using sugar beet pulp. Process Biochem. 2011, 46, 1076–1082. [Google Scholar] [CrossRef]
- Akintelu, S.A.; Yao, B.; Folorunso, A.S. Green Synthesis, Characterization, and Antibacterial Investigation of Synthesized Gold Nanoparticles (AuNPs) from Garcinia kola Pulp Extract. Plasmonics 2020, 1–9. [Google Scholar] [CrossRef]
- Wongyai, K.; Wintachai, P.; Maungchang, R.; Rattanakit, P. Exploration of the Antimicrobial and Catalytic Properties of Gold Nanoparticles Greenly Synthesized by Cryptolepis buchanani Roem. And Schult Extract. J. Nanomater. 2020. [Google Scholar] [CrossRef]
- Shah, A.; Latif-Ur-Rahman; Qureshi, R.; Zia-Ur-Rehman. Synthesis, characterization and applications of bimetallic (Au-Ag, Au-Pt, Au-Ru) alloy nanoparticles. Rev. Adv. Mater. Sci. 2012, 30, 133–149. [Google Scholar]
- Radziuk, D.V.; Zhang, W.; Shchukin, D.; Möhwald, H. Ultrasonic alloying of preformed gold and silver nanoparticles. Small 2010, 6, 545–553. [Google Scholar] [CrossRef]
- Duan, S.; Wang, R. Bimetallic nanostructures with magnetic and noble metals and their physicochemical applications. Prog. Nat. Sci. Mater. Int. 2013, 23, 113–126. [Google Scholar] [CrossRef] [Green Version]
- Elemike, E.E.; Onwudiwe, D.C.; Fayemi, O.E.; Botha, T.L. Green synthesis and electrochemistry of Ag, Au, and Ag–Au bimetallic nanoparticles using golden rod (Solidago canadensis) leaf extract. Appl. Phys. A Mater. Sci. Process. 2019, 125, 42. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Wang, A.; He, Q.; Li, J. Hierarchical gold/copolymer nanostructures as hydrophobic nanotanks for drug encapsulation. J. Mater. Chem. 2010, 20, 7782–7787. [Google Scholar] [CrossRef]
- Elemike, E.E.; Onwudiwe, D.C.; Nundkumar, N.; Singh, M.; Iyekowa, O. Green synthesis of Ag, Au and Ag-Au bimetallic nanoparticles using Stigmaphyllon ovatum leaf extract and their in vitro anticancer potential. Mater. Lett. 2019, 243, 148–152. [Google Scholar] [CrossRef]
- Gopinath, K.; Kumaraguru, S.; Bhakyaraj, K.; Mohan, S.; Venkatesh, K.S.; Esakkirajan, M.; Kaleeswarran, P.; Alharbi, N.S.; Kadaikunnan, S.; Govindarajan, M.; et al. Green synthesis of silver, gold and silver/gold bimetallic nanoparticles using the Gloriosa superba leaf extract and their antibacterial and antibiofilm activities. Microb. Pathog. 2016, 101, 1–11. [Google Scholar] [CrossRef]
- Kumari, M.M.; Jacob, J.; Philip, D. Green synthesis and applications of Au–Ag bimetallic nanoparticles. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015, 137, 185–192. [Google Scholar] [CrossRef]
- Emam, H.E. Arabic Gum as Bio-Synthesizer for Ag–Au Bimetallic Nanocomposite Using Seed-Mediated Growth Technique and Its Biological Efficacy. J. Polym. Environ. 2019, 27, 210–223. [Google Scholar] [CrossRef]
- Gupta, S.; Hemlata, H.; Tejavath, K. Synthesis, characterization and comparative anticancer potential of phytosynthesized mono and bimetallic nanoparticles using Moringa oleifera aqueous leaf extract. Beilstein Arch. 2020, 1, 95. [Google Scholar]
- Li, X.-Z. Antimicrobial Resistance in Bacteria: An Overview of Mechanisms and Role of Drug Efflux Pumps. In Efflux-Mediated Antimicrobial Resistance in Bacteria; Adis: Cham, Switzerland, 2016. [Google Scholar]
- Blair, J.M.A.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J.V. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Hajipour, M.J.; Fromm, K.M.; Ashkarran, A.A.; de Aberasturi, D.J.; de Larramendi, I.R.; Rojo, T.; Serpooshan, V.; Parak, W.J.; Mahmoudi, M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012, 30, 499–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaraman, R. Antibiotic resistance: An overview of mechanisms and a paradigm shift. Curr. Sci. 2009, 96, 1475–1484. [Google Scholar]
- Barlow, M. What antimicrobial resistance has taught us about horizontal gene transfer. In Horizontal Gene Transfer; Humana Press: Totowa, NJ, USA, 2009. [Google Scholar]
- Todar, K. Todar’s Online Textbook of Bacteriology; University of Wisconsin-Madison Department of Bacteriology: Madison, WI, USA, 2011. [Google Scholar]
- Sommer, M.O.A.; Munck, C.; Toft-Kehler, R.V.; Andersson, D.I. Prediction of antibiotic resistance: Time for a new preclinical paradigm? Nat. Rev. Microbiol. 2017, 15, 689–696. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, M.C.; Sanchez-Lopez, E.; Espina, M.; Calpena, A.C.; Silva, A.M.; Veiga, F.J.; Garcia, M.L.; Souto, E.B. Chapter 9—Advances in antibiotic nanotherapy: Overcoming antimicrobial resistance. In Emerging Nanotechnologies in Immunology; Elsevier: Amsterdam, The Netherlands, 2018. [Google Scholar]
- Lara, H.H.; Ayala-Núñez, N.V.; del Turrent, L.C.I.; Padilla, C.R. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J. Microbiol. Biotechnol. 2010, 26, 615–621. [Google Scholar] [CrossRef]
- Poole, K. Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 2002, 92, 55S–64S. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Y.; Jin, M.; Yuan, Z.; Bond, P.; Guo, J. Both silver ions and silver nanoparticles facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes. Water Res. 2020, 169, 115229. [Google Scholar] [CrossRef] [PubMed]
- Graves, J.L.; Tajkarimi, M.; Cunningham, Q.; Campbell, A.; Nonga, H.; Harrison, S.H.; Barrick, J.E. Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front. Genet. 2015, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.L.; Hudson-Smith, N.V.; Cahill, M.S.; Reynolds, B.N.; Frand, S.D.; Green, C.M.; Wang, C.; Hang, M.N.; Hernandez, R.T.; Hamers, R.J.; et al. Chronic exposure to complex metal oxide nanoparticles elicits rapid resistance in: Shewanella oneidensis MR-1. Chem. Sci. 2019, 10, 9768–9781. [Google Scholar] [CrossRef] [Green Version]
- Salas Orozco, M.F.; Niño-Martínez, N.; Martínez-Castañón, G.A.; Méndez, F.T.; Ruiz, F. Molecular mechanisms of bacterial resistance to metal and metal oxide nanoparticles. Int. J. Mol. Sci. 2019, 20, 2808. [Google Scholar]
- Panáček, A.; Kvítek, L.; Smékalová, M.; Večeřová, R.; Kolář, M.; Röderová, M.; Dyčka, F.; Šebela, M.; Prucek, R.; Tomanec, O.; et al. Bacterial resistance to silver nanoparticles and how to overcome it. Nat. Nanotechnol. 2018, 13, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Siemer, S.; Westmeier, D.; Barz, M.; Eckrich, J.; Wünsch, D.; Seckert, C.; Thyssen, C.; Schilling, O.; Hasenberg, M.; Pang, C.; et al. Biomolecule-corona formation confers resistance of bacteria to nanoparticle-induced killing: Implications for the design of improved nanoantibiotics. Biomaterials 2019, 192, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Kumariya, R.; Sood, S.K.; Rajput, Y.S.; Saini, N.; Garsa, A.K. Increased membrane surface positive charge and altered membrane fluidity leads to cationic antimicrobial peptide resistance in Enterococcus faecalis. Biochim. Biophys. Acta Biomembr. 2015, 1848, 1367–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Sun, R.; Xia, T. Adaption/resistance to antimicrobial nanoparticles: Will it be a problem? Nano Today 2020, 34, 100909. [Google Scholar] [CrossRef]
- Kittler, S.; Greulich, C.; Diendorf, J.; Köller, M.; Epple, M. Toxicity of silver nanoparticles increases during storage because of slow dissolution under release of silver ions. Chem. Mater. 2010, 22, 4548–4554. [Google Scholar] [CrossRef]
- Feng, Q.L.; Wu, J.; Chen, G.Q.; Cui, F.Z.; Kim, T.N.; Kim, J.O. A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus. J. Biomed. Mater. Res. 2000, 52, 662–668. [Google Scholar] [CrossRef]
- Kim, T.H.; Kim, M.; Park, H.S.; Shin, U.S.; Gong, M.S.; Kim, H.W. Size-dependent cellular toxicity of silver nanoparticles. J. Biomed. Mater. Res. Part. A 2012, 100, 1033–1043. [Google Scholar] [CrossRef]
- Mukha, I.P.; Eremenko, A.M.; Smirnova, N.P.; Mikhienkova, A.I.; Korchak, G.I.; Gorchev, V.F.; Chunikhin, A.Y. Antimicrobial activity of stable silver nanoparticles of a certain size. Appl. Biochem. Microbiol. 2013, 49, 199–206. [Google Scholar] [CrossRef]
- Cavassin, E.D.; de Figueiredo, L.F.P.; Otoch, J.P.; Seckler, M.M.; de Oliveira, R.A.; Franco, F.F.; Marangoni, V.S.; Zucolotto, V.; Levin, A.S.S.; Costa, S.F. Comparison of methods to detect the in vitro activity of silver nanoparticles (AgNP) against multidrug resistant bacteria. J. Nanobiotechnol. 2015, 13, 64. [Google Scholar] [CrossRef]
- Dorobantu, L.S.; Fallone, C.; Noble, A.J.; Veinot, J.; Ma, G.; Goss, G.G.; Burrell, R.E. Toxicity of silver nanoparticles against bacteria, yeast, and algae. J. Nanopart. Res. 2015, 17, 172. [Google Scholar] [CrossRef]
- Edis, Z.; Bloukh, S.H.; Ibrahim, M.R.; Sara, H.A. “Smart” Antimicrobial Nanocomplexes with Potential to Decrease Surgical Site Infections (SSI). Pharmaceutics 2020, 12, 361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnusson, K.E.; Bayer, M.E. Anionic sites on the envelope of Salmonella typhimurium mapped with cationized ferritin. Cell Biophys. 1982, 4, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Sonohara, R.; Muramatsu, N.; Ohshima, H.; Kondo, T. Difference in surface properties between Escherichia coli and Staphylococcus aureus as revealed by electrophoretic mobility measurements. Biophys. Chem. 1995, 55, 273–277. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Holt, K.B.; Bard, A.J. Interaction of silver(I) ions with the respiratory chain of Escherichia coli: An electrochemical and scanning electrochemical microscopy study of the antimicrobial mechanism of micromolar Ag. Biochemistry 2005, 44, 13214–13223. [Google Scholar] [CrossRef]
- Durán, N.; Durán, M.; de Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 789–799. [Google Scholar] [CrossRef]
- Alsammarraie, F.K.; Wang, W.; Zhou, P.; Mustapha, A.; Lin, M. Green synthesis of silver nanoparticles using turmeric extracts and investigation of their antibacterial activities. Colloids Surf. B Biointerfaces 2018, 171, 398–405. [Google Scholar] [CrossRef]
- Rattanata, N.; Klaynongsruang, S.; Leelayuwat, C.; Limpaiboon, T.; Lulitanond, A.; Boonsiri, P.; Chio-Srichan, S.; Soontaranon, S.; Rugmai, S.; Daduang, J. Gallic acid conjugated with gold nanoparticles: Antibacterial activity and mechanism of action on foodborne pathogens. Int. J. Nanomed. 2016, 11, 3347–3356. [Google Scholar] [CrossRef] [Green Version]
- Burgula, Y.; Khali, D.; Kim, S.; Krishnan, S.S.; Cousin, M.A.; Gore, J.P.; Reuhs, B.L.; Mauer, L.J. Review of mid-infrared fourier transform-infrared spectroscopy applications for bacterial detection. J. Rapid Methods Autom. Microbiol. 2007, 15, 146–175. [Google Scholar] [CrossRef]
- Riding, M.J.; Martin, F.L.; Trevisan, J.; Llabjani, V.; Patel, I.I.; Jones, K.C.; Semple, K.T. Concentration-dependent effects of carbon nanoparticles in gram-negative bacteria determined by infrared spectroscopy with multivariate analysis. Environ. Pollut. 2012, 163, 226–234. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Mouwen, D.J.M.; López, M.; Prieto, M. Fourier transform infrared spectroscopy as a tool to characterize molecular composition and stress response in foodborne pathogenic bacteria. J. Microbiol. Methods 2011, 84, 369–378. [Google Scholar] [CrossRef] [PubMed]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free Radicals: Properties, Sources, Targets, and Their Implication in Various Diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef] [Green Version]
- Ravichandran, A.; Subramanian, P.; Manoharan, V.; Muthu, T.; Periyannan, R.; Thangapandi, M.; Ponnuchamy, K.; Pandi, B.; Marimuthu, P.N. Phyto-mediated synthesis of silver nanoparticles using fucoidan isolated from Spatoglossum asperum and assessment of antibacterial activities. J. Photochem. Photobiol. B Biol. 2018, 185, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Jha, E.; Sahoo, B.; Panda, P.K.; Thirumurugan, A.; Parashar, S.K.S.; Suar, M. Mechanistic insight into the rapid one-step facile biofabrication of antibacterial silver nanoparticles from bacterial release and their biogenicity and concentration-dependent in vitro cytotoxicity to colon cells. RSC Adv. 2017, 7, 40034–40045. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Yuan, J.; Gao, H. Microbial oxidative stress response: Novel insights from environmental facultative anaerobic bacteria. Arch. Biochem. Biophys. 2015, 584, 28–35. [Google Scholar] [CrossRef]
- Zhang, Y.; Meng, D.; Wang, Z.; Guo, H.; Wang, Y. Oxidative stress response in two representative bacteria exposed to atrazine. FEMS Microbiol. Lett. 2012, 334, 95–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abd-Elnaby, H.M.; Abo-Elala, G.M.; Abdel-Raouf, U.M.; Hamed, M.M. Antibacterial and anticancer activity of extracellular synthesized silver nanoparticles from marine Streptomyces rochei MHM13. Egypt. J. Aquat. Res. 2016, 42, 301–312. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.S.; Kuk, E.; Yu, K.N.; Kim, J.H.; Park, S.J.; Lee, H.J.; Kim, S.H.; Park, Y.K.; Park, Y.H.; Hwang, C.Y.; et al. Antimicrobial effects of silver nanoparticles. Nanomed. Nanotechnol. Biol. Med. 2007, 3, 95–101. [Google Scholar] [CrossRef]
- Chakraborty, S.; Chelli, V.R.; Das, R.K.; Giri, A.S.; Golder, A.K. Bio-mediated silver nanoparticle synthesis: Mechanism and microbial inactivation. Toxicol. Environ. Chem. 2017, 99, 434–447. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, H.S.; Ryu, D.S.; Choi, S.J.; Lee, D.S. Antibacterial activity of silver-nanoparticles against Staphylococcus aureus and Escherichia coli. Korean J. Microbiol. Biotechnol. 2011, 39, 77–85. [Google Scholar]
- Gomaa, E.Z. Silver nanoparticles as an antimicrobial agent: A case study on Staphylococcus aureus and escherichia coli as models for gram-positive and gram-negative bacteria. J. Gen. Appl. Microbiol. 2017, 63, 36–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qayyum, S.; Oves, M.; Khan, A.U. Obliteration of bacterial growth and biofilm through ROS generation by facilely synthesized green silver nanoparticles. PLoS ONE 2017, 12, e0181363. [Google Scholar] [CrossRef] [Green Version]
- Farah, M.A.; Ali, M.A.; Chen, S.M.; Li, Y.; Al-Hemaid, F.M.; Abou-Tarboush, F.M.; Al-Anazi, K.M.; Lee, J. Silver nanoparticles synthesized from Adenium obesum leaf extract induced DNA damage, apoptosis and autophagy via generation of reactive oxygen species. Colloids Surf. B Biointerfaces 2016, 141, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Qu, F.; Xu, H.; Lai, W.; Wang, Y.A.; Aguilar, Z.P.; Wei, H. Role of reactive oxygen species in the antibacterial mechanism of silver nanoparticles on Escherichia coli O157:H7. BioMetals 2012, 25, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.; Zhang, Y.; Pan, X.; Zhu, F.; Jiang, C.; Liu, Q.; Cheng, Z.; Dai, G.; Wu, G.; Wang, L.; et al. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant pseudomonas aeruginosa. Int. J. Nanomed. 2019, 14, 1469–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y.; Zhao, Y.; Tian, Y.; Zhang, W.; Lü, X.; Jiang, X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 2012, 33, 2327–2333. [Google Scholar] [CrossRef]
- Abbas, W.S.; Atwan, Z.W.; Abdulhussein, Z.R.; Mahdi, M.A. Preparation of silver nanoparticles as antibacterial agents through DNA damage. Mater. Technol. 2019, 34, 867–879. [Google Scholar] [CrossRef]
- Mousavi-Khattat, M.; Keyhanfar, M.; Razmjou, A. A comparative study of stability, antioxidant, DNA cleavage and antibacterial activities of green and chemically synthesized silver nanoparticles. Artif. Cells Nanomed. Biotechnol. 2018, 46, S1022–S1031. [Google Scholar] [CrossRef]
- Gulbagca, F.; Ozdemir, S.; Gulcan, M.; Sen, F. Synthesis and characterization of Rosa canina-mediated biogenic silver nanoparticles for anti-oxidant, antibacterial, antifungal, and DNA cleavage activities. Heliyon 2019, 5, e02980. [Google Scholar] [CrossRef] [Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fanoro, O.T.; Oluwafemi, O.S. Bactericidal Antibacterial Mechanism of Plant Synthesized Silver, Gold and Bimetallic Nanoparticles. Pharmaceutics 2020, 12, 1044. https://doi.org/10.3390/pharmaceutics12111044
Fanoro OT, Oluwafemi OS. Bactericidal Antibacterial Mechanism of Plant Synthesized Silver, Gold and Bimetallic Nanoparticles. Pharmaceutics. 2020; 12(11):1044. https://doi.org/10.3390/pharmaceutics12111044
Chicago/Turabian StyleFanoro, Olufunto T., and Oluwatobi S. Oluwafemi. 2020. "Bactericidal Antibacterial Mechanism of Plant Synthesized Silver, Gold and Bimetallic Nanoparticles" Pharmaceutics 12, no. 11: 1044. https://doi.org/10.3390/pharmaceutics12111044