Design and Characterization of Glyceryl Monooleate-Nanostructures Containing Doxorubicin Hydrochloride
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
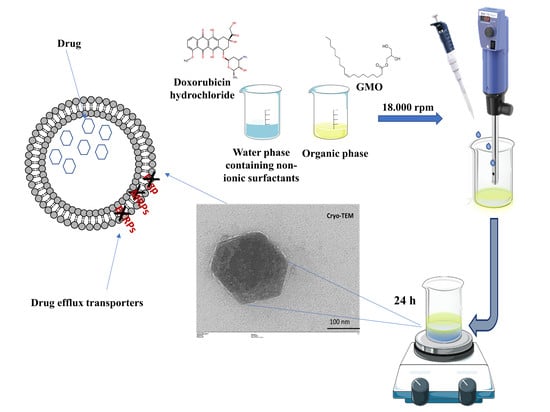
2.2. Preparation of GMO-Nanostructures
2.3. Physico-Chemical Characterization
2.3.1. Influence of Temperature, pH and Serum on the Stability of Nanostructures
2.3.2. Freeze-Drying of Nanosystems
2.4. Entrapment Efficiency of DOX
2.5. DOX Release from GMO-Nanostructures
2.6. Cell Cultures and In Vitro Cytotoxicity
Cytotoxicity of DOX-Loaded Nanostructures with Verapamil
2.7. Interaction of Tritiated GMO-Nanostructures with Cells
2.8. Cell Interaction by CLSM
2.9. Statistical Analysis
3. Results and Discussion
3.1. Physico-Chemical and Technological Characterization of Nanostructures
3.2. Physico-Chemical Characterization of DOX-Loaded GMO-Nanostructures
Evaluation of the Entrapment Efficiency and Release Profiles of DOX
3.3. Cytotoxicity of GMO-Nanostructures
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fong, C.; Le, T.; Drummond, C.J. Lyotropic liquid crystal engineering–ordered nanostructured small molecule amphiphile self-assembly materials by design. Chem. Soc. Rev. 2012, 41, 1297–1322. [Google Scholar] [CrossRef] [PubMed]
- Urandur, S.; Marwaha, D.; Gautam, S.; Banala, V.T.; Sharma, M.; Mishra, P.R. Nonlamellar liquid crystals: A new paradigm for the delivery of small molecules and bio-macromolecules. Ther. Deliv. 2018, 9, 667–689. [Google Scholar] [CrossRef] [PubMed]
- Akbar, S.; Anwar, A.; Ayish, A.; Elliott, J.M.; Squires, A.M. Phytantriol based smart nano-carriers for drug delivery applications. Eur. J. Pharm. Sci. 2017, 101, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Bode, J.C.; Kuntsche, J.; Funari, S.S.; Bunjes, H. Interaction of dispersed cubic phases with blood components. Int. J. Pharm. 2013, 448, 87–95. [Google Scholar] [CrossRef]
- Matschke, C.; Isele, U.; Van Hoogevest, P.; Fahr, A. Sustained-release injectables formed in situ and their potential use for veterinary products. J. Control Release 2002, 85, 1–15. [Google Scholar] [CrossRef]
- Paolino, D.; Tudose, A.; Celia, C.; Di Marzio, L.; Cilurzo, F.; Mircioiu, C. Mathematical Models as Tools to Predict the Release Kinetic of Fluorescein from Lyotropic Colloidal Liquid Crystals. Materials 2019, 12, 693. [Google Scholar] [CrossRef] [Green Version]
- Karami, Z.; Hamidi, M. Cubosomes: Remarkable drug delivery potential. Drug Discov. Today 2016, 21, 789–801. [Google Scholar] [CrossRef]
- Barriga, H.M.G.; Holme, M.N.; Stevens, M.M. Cubosomes: The Next Generation of Smart Lipid Nanoparticles? Angew. Chem. Int. Ed. Engl. 2019, 58, 2958–2978. [Google Scholar] [CrossRef] [Green Version]
- Hirlekar, R.; Jain, S.; Patel, M.; Garse, H.; Kadam, V. Hexosomes: A novel drug delivery system. Curr. Drug Deliv. 2010, 7, 28–35. [Google Scholar] [CrossRef]
- Rodrigues, L.; Kyriakos, K.; Schneider, F.; Dietz, H.; Winter, G.; Papadakis, C.M.; Hubert, M. Characterization of Lipid-Based Hexosomes as Versatile Vaccine Carriers. Mol. Pharm. 2016, 13, 3945–3954. [Google Scholar] [CrossRef]
- Rodrigues, L.; Schneider, F.; Zhang, X.; Larsson, E.; Moodie, L.W.K.; Dietz, H.; Papadakis, C.M.; Winter, G.; Lundmark, R.; Hubert, M. Cellular uptake of self-assembled phytantriol-based hexosomes is independent of major endocytic machineries. J. Colloid Interface Sci. 2019, 553, 820–833. [Google Scholar] [CrossRef] [PubMed]
- Murgia, S.; Biffi, S.; Mezzenga, R. Recent advances of non-lamellar lyotropic liquid crystalline nanoparticles in nanomedicine. Curr. Opin. Colloid Interface Sci. 2020, 48, 28–39. [Google Scholar] [CrossRef]
- Mertins, O.; Mathews, P.D.; Angelova, A. Advances in the Design of pH-Sensitive Cubosome Liquid Crystalline Nanocarriers for Drug Delivery Applications. Nanomaterials 2020, 10, 963. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, E.; Paolino, D.; Fresta, M.; Cosco, D. Mucosal applications of poloxamer 407-based hydrogels: An overview. Pharmaceutics 2018, 10, 159. [Google Scholar] [CrossRef] [Green Version]
- Giuliano, E.; Paolino, D.; Fresta, M.; Cosco, D. Drug-Loaded Biocompatible Nanocarriers Embedded in Poloxamer 407 Hydrogels as Therapeutic Formulations. Medicines 2018, 6, 7. [Google Scholar] [CrossRef] [Green Version]
- Amar-Yuli, I.; Wachtel, E.; Shoshan, E.B.; Danino, D.; Aserin, A.; Garti, N. Hexosome and hexagonal phases mediated by hydration and polymeric stabilizer. Langmuir 2007, 23, 3637–3645. [Google Scholar] [CrossRef]
- Khaliq, N.U.; Park, D.Y.; Yun, B.M.; Jung, Y.W.; Seo, J.H.; Hwang, C.S.; Yuk, S.H. Pluronics: Intelligent building units for targeted cancer therapy and molecular imaging. Int. J. Pharm. 2019, 556, 30–44. [Google Scholar] [CrossRef]
- Kerwin, B.A. Polysorbates 20 and 80 used in the formulation of protein biotherapeutics: Structure and degradation pathways. J. Pharm. Sci. 2008, 97, 2924–2935. [Google Scholar] [CrossRef]
- Minotti, G.; Recalcati, S.; Menna, P.; Salvatorelli, E.; Corna, G.; Cairo, G. Doxorubicin cardiotoxicity and the control of iron metabolism: Quinone-dependent and independent mechanisms. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2004; Volume 378, pp. 340–361. ISBN 0076-6879. [Google Scholar]
- Tasca, E.; Del Giudice, A.; Galantini, L.; Schillén, K.; Giuliani, A.M.; Giustini, M. A fluorescence study of the loading and time stability of doxorubicin in sodium cholate/PEO-PPO-PEO triblock copolymer mixed micelles. J. Colloid Interface Sci. 2019, 540, 593–601. [Google Scholar] [CrossRef]
- Nazaruk, E.; Szlęzak, M.; Górecka, E.; Bilewicz, R.; Osornio, Y.M.; Uebelhart, P.; Landau, E.M. Design and assembly of pH-sensitive lipidic cubic phase matrices for drug release. Langmuir 2014, 30, 1383–1390. [Google Scholar] [CrossRef]
- Szakács, G.; Paterson, J.K.; Ludwig, J.A.; Booth-Genthe, C.; Gottesman, M.M. Targeting multidrug resistance in cancer. Nat. Rev. Drug Discov. 2006, 5, 219. [Google Scholar] [CrossRef] [PubMed]
- Muheem, A.; Shakeel, F.; Warsi, M.H.; Jain, G.K.; Ahmad, F.J. A Combinatorial Statistical Design Approach to Optimize the Nanostructured Cubosomal Carrier System for Oral Delivery of Ubidecarenone for Management of Doxorubicin-Induced Cardiotoxicity: In Vitro-In Vivo Investigations. J. Pharm. Sci. 2017, 106, 3050–3065. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Angelova, A.; Hu, F.; Garamus, V.M.; Peng, C.; Li, N.; Liu, J.; Liu, D.; Zou, A. pH Responsiveness of Hexosomes and Cubosomes for Combined Delivery of Brucea javanica Oil and Doxorubicin. Langmuir 2019, 35, 14532–14542. [Google Scholar] [CrossRef] [PubMed]
- Barenholz, Y.C. Doxil®—The first FDA-approved nano-drug: Lessons learned. J. Control Release 2012, 160, 117–134. [Google Scholar] [CrossRef]
- Cosco, D.; Federico, C.; Maiuolo, J.; Bulotta, S.; Molinaro, R.; Paolino, D.; Tassone, P.; Fresta, M. Physicochemical features and transfection properties of chitosan/poloxamer 188/poly(D,L-lactide-co-glycolide) nanoplexes. Int. J. Nanomed. 2014, 9, 2359–2372. [Google Scholar] [CrossRef] [Green Version]
- Cosco, D.; Paolino, D.; De Angelis, F.; Cilurzo, F.; Celia, C.; Di Marzio, L.; Russo, D.; Tsapis, N.; Fattal, E.; Fresta, M. Aqueous-core PEG-coated PLA nanocapsules for an efficient entrapment of water soluble anticancer drugs and a smart therapeutic response. Eur. J. Pharm. Biopharm. 2015, 89, 30–39. [Google Scholar] [CrossRef]
- Gagliardi, A.; Froiio, F.; Salvatici, M.C.; Paolino, D.; Fresta, M.; Cosco, D. Characterization and refinement of zein-based gels. Food Hydrocoll. 2020, 101, 105555. [Google Scholar] [CrossRef]
- Sun, C.; Wu, T.; Liu, R.; Liang, B.; Tian, Z.; Zhang, E.; Zhang, M. Effects of superfine grinding and microparticulation on the surface hydrophobicity of whey protein concentrate and its relation to emulsions stability. Food Hydrocoll. 2015, 51, 512–518. [Google Scholar] [CrossRef]
- Gagliardi, A.; Bonacci, S.; Paolino, D.; Celia, C.; Procopio, A.; Fresta, M.; Cosco, D. Paclitaxel-loaded sodium deoxycholate-stabilized zein nanoparticles: Characterization and in vitro cytotoxicity. Heliyon 2019, 5, e02422. [Google Scholar] [CrossRef] [Green Version]
- Cosco, D.; Tsapis, N.; Nascimento, T.L.; Fresta, M.; Chapron, D.; Taverna, M.; Arpicco, S.; Fattal, E. Polysaccharide-coated liposomes by post-insertion of a hyaluronan-lipid conjugate. Colloids Surf. B Biointerfaces 2017, 158, 119–126. [Google Scholar] [CrossRef] [Green Version]
- Gagliardi, A.; Paolino, D.; Iannone, M.; Palma, E.; Fresta, M.; Cosco, D. Sodium deoxycholate-decorated zein nanoparticles for a stable colloidal drug delivery system. Int. J. Nanomed. 2018, 13, 601–614. [Google Scholar] [CrossRef] [Green Version]
- Gagliardi, A.; Voci, S.; Paolino, D.; Fresta, M.; Cosco, D. Influence of Various Model Compounds on the Rheological Properties of Zein-Based Gels. Molecules 2020, 25, 3174. [Google Scholar] [CrossRef]
- Cosco, D.; Mare, R.; Paolino, D.; Salvatici, M.C.; Cilurzo, F.; Fresta, M. Sclareol-loaded hyaluronan-coated PLGA nanoparticles: Physico-chemical properties and in vitro anticancer features. Int. J. Biol. Macromol. 2019, 132, 550–557. [Google Scholar] [CrossRef]
- Bulotta, S.; Corradino, R.; Celano, M.; D’Agostino, M.; Maiuolo, J.; Oliverio, M.; Procopio, A.; Iannone, M.; Rotiroti, D.; Russo, D. Antiproliferative and antioxidant effects on breast cancer cells of oleuropein and its semisynthetic peracetylated derivatives. Food Chem. 2011, 127, 1609–1614. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, D.; Zhang, Q.; Chen, Y.; Zheng, D.; Hao, L.; Duan, C.; Jia, L.; Liu, G.; Liu, Y. Synergistic effect of folate-mediated targeting and verapamil-mediated P-gp inhibition with paclitaxel-polymer micelles to overcome multi-drug resistance. Biomaterials 2011, 32, 9444–9456. [Google Scholar] [CrossRef]
- Bao, L.; Hazari, S.; Mehra, S.; Kaushal, D.; Moroz, K.; Dash, S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am. J. Pathol. 2012, 180, 2490–2503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.Y.T.; Mulet, X.; Waddington, L.J.; Boyd, B.J.; Drummond, C.J. Steric stabilisation of self-assembled cubic lyotropic liquid crystalline nanoparticles: High throughput evaluation of triblock polyethylene oxide-polypropylene oxide-polyethylene oxide copolymers. Soft Matter 2011, 7, 4768–4777. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential—What they are and what they are not? J. Control Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Molinaro, R.; Gagliardi, A.; Mancuso, A.; Cosco, D.; Soliman, M.E.; Casettari, L.; Paolino, D. Development and In Vivo Evaluation of Multidrug Ultradeformable Vesicles for the Treatment of Skin Inflammation. Pharmaceutics 2019, 11, 644. [Google Scholar] [CrossRef] [Green Version]
- Giuliano, E.; Paolino, D.; Cristiano, M.C.; Fresta, M.; Cosco, D. Rutin-Loaded Poloxamer 407-Based Hydrogels for In Situ Administration: Stability Profiles and Rheological Properties. Nanomaterials 2020, 10, 1069. [Google Scholar] [CrossRef]
- Gagliardi, A.; Paolino, D.; Costa, N.; Fresta, M.; Cosco, D. Zein-vs PLGA-based nanoparticles containing rutin: A comparative investigation. Mater. Sci. Eng. C 2020, 118, 111538. [Google Scholar] [CrossRef]
- Gupta, U.; Sharma, S.; Khan, I.; Gothwal, A.; Sharma, A.K.; Singh, Y.; Chourasia, M.K.; Kumar, V. Enhanced apoptotic and antican cer potential of paclitaxel loaded biodegradable nanoparticles based on chitosan. Int. J. Biol. Macromol. 2017, 98, 810–819. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.; Das, S.; Mondal, A.; Chatterji, U.; Mukherjee, A. Nanoparticle engineering enhances anticancer efficacy of andrographolide in MCF-7 cells and mice bearing EAC. Curr. Pharm. Biotechnol. 2012, 13, 2669–2681. [Google Scholar] [CrossRef] [PubMed]
- Batrakova, E.V.; Li, S.; Vinogradov, S.V.; Alakhov, V.Y.; Miller, D.W.; Kabanov, A.V. Mechanism of pluronic effect on P-glycoprotein efflux system in blood-brain barrier: Contributions of energy depletion and membrane fluidization. J. Pharmacol. Exp. Ther. 2001, 299, 483–493. [Google Scholar]
- Alakhova, D.Y.; Kabanov, A.V. Pluronics and MDR reversal: An update. Mol. Pharm. 2014, 11, 2566–2578. [Google Scholar] [CrossRef]
- Sobot, D.; Mura, S.; Couvreur, P. How can nanomedicines overcome cellular-based anticancer drug resistance? J. Mater. Chem. B 2016, 4, 5078–5100. [Google Scholar] [CrossRef]
- Alvarez-Lorenzo, C.; Sosnik, A.; Concheiro, A. PEO-PPO block copolymers for passive micellar targeting and overcoming multidrug resistance in cancer therapy. Curr. Drug Targets 2011, 12, 1112–1130. [Google Scholar] [CrossRef]








| Sample | Molecular Weight | HLB | Mean Sizes (nm) | Polydispersity Index | Zeta Potential (mV) |
|---|---|---|---|---|---|
| PL F-127 a | 12,500 | 18–23 | 96 ± 2 | 0.11 ± 0.02 | −27 ± 1 |
| PL F-68 b | 8500 | 29 | 229 ± 1 | 0.19 ± 0.03 | −17 ± 2 |
| PL 10500 c | 6500 | 15 | 327 ± 2 | 0.34 ± 0.05 | −20 ± 2 |
| T20 d | 1227 | 16.7 | 108 ± 1 | 0.09 ± 0.01 | −27 ± 1 |
| T40 e | 1284 | 15.6 | 105 ± 2 | 0.31 ± 0.02 | −29 ± 1 |
| T60 f | 1311 | 14.9 | 93 ± 1 | 0.11 ± 0.03 | −26 ± 2 |
| T80 g | 1310 | 15 | 120 ± 2 | 0.07 ± 0.05 | −26 ± 2 |
| Sample | Temperature (°C) | Mean Size (nm) | Polydispersity Index | Zeta Potential (mV) |
|---|---|---|---|---|
| PL F127 | Before heating | 96 ± 2 | 0.11 ± 0.02 | −27 ± 1 |
| 30 | 114 ± 6 | 0.28 ± 0.02 | −22 ± 1 | |
| 40 | 114 ± 6 | 0.28 ± 0.07 | −21 ± 1 | |
| 50 | 118 ± 6 | 0.31 ± 0.08 | −20 ± 2 | |
| PL F68 | Before heating | 229 ± 1 | 0.19 ± 0.03 | −17 ± 2 |
| 30 | 220 ± 8 | 0.32 ± 0.01 | −23 ± 1 | |
| 40 | 224 ± 8 | 0.35 ± 0.01 | −23 ± 1 | |
| 50 | 242 ± 8 | 0.31 ± 0.01 | −22 ± 1 | |
| PL 10500 | Before heating | 327 ± 2 | 0.34 ± 0.05 | −20 ± 2 |
| 30 | 381 ± 9 | 0.32 ± 0.03 | −22 ± 1 | |
| 40 | 354 ± 9 | 0.33 ± 0.02 | −23 ± 1 | |
| 50 | 389 ± 9 | 0.36 ± 0.01 | −23 ± 1 | |
| T20 | Before heating | 108 ± 1 | 0.09 ± 0.01 | −27 ± 1 |
| 30 | 112 ± 5 | 0.16 ± 0.01 | −26 ± 2 | |
| 40 | 120 ± 6 | 0.26 ± 0.02 | −25 ± 2 | |
| 50 | 115 ± 5 | 0.11 ± 0.02 | −24 ± 1 | |
| T40 | Before heating | 105 ± 2 | 0.31 ± 0.02 | −29 ± 1 |
| 30 | 72 ± 3 | 0.24 ± 0.01 | −30 ± 3 | |
| 40 | 76 ± 4 | 0.20 ± 0.02 | −25 ± 2 | |
| 50 | 72 ± 4 | 0.20 ± 0.02 | −26 ± 1 | |
| T60 | Before heating | 93 ± 1 | 0.11 ± 0.03 | −26 ± 2 |
| 30 | 96 ± 1 | 0.10 ± 0.01 | −26 ± 2 | |
| 40 | 96 ± 2 | 0.11 ± 0.02 | −20 ± 2 | |
| 50 | 101 ± 1 | 0.10 ± 0.02 | −20 ± 1 | |
| T80 | Before heating | 120 ± 2 | 0.07 ± 0.05 | −26 ± 2 |
| 30 | 126 ± 6 | 0.13 ± 0.01 | −27 ± 3 | |
| 40 | 132 ± 7 | 0.22 ± 0.02 | −28 ± 2 | |
| 50 | 128 ± 6 | 0.13 ± 0.02 | −26 ± 1 |
| Nano F-127 | |||
|---|---|---|---|
| DOX Concentration (mg/mL) | Mean Sizes (nm) | Polydispersity Index | Zeta Potential (mV) |
| 0.1 | 94 ± 1 | 0.19 ± 0.02 | −17 ± 1 |
| 0.2 | 97 ± 5 | 0.14 ± 0.04 | −20 ± 1 |
| 0.4 | 112 ± 6 | 0.18 ± 0.08 | −22 ± 2 |
| 0.6 | 122 ± 7 | 0.22 ± 0.04 | −22 ± 2 |
| 0.8 | 280 ± 5 | 0.47 ± 0.02 | −16 ± 2 |
| Nano T80 | |||
| 0.1 | 120 ± 1 | 0.12 ± 0.02 | −24 ± 1 |
| 0.2 | 132 ± 4 | 0.19 ± 0.04 | −22 ± 1 |
| 0.4 | 150 ± 6 | 0.21 ± 0.08 | −25 ± 2 |
| 0.6 | 185 ± 7 | 0.28 ± 0.04 | −27 ± 2 |
| 0.8 | 296 ± 5 | 0.5 ± 0.06 | −20 ± 2 |
| Cell Lines | Incubation Time | IC50 DOX | IC50 T80 DOX | IC50 PL F127 DOX |
|---|---|---|---|---|
| MCF-7 | 24 h | 10.16 | 8.71 | 1.71 |
| 48 h | 9.79 | 5.18 | 0.66 | |
| 72 h | 0.75 | 0.52 | 0.28 | |
| MDA-MB-231 | 24 h | 9.41 | 3.72 | 0.86 |
| 48 h | 2.86 | 0.79 | 0.32 | |
| 72 h | 0.46 | 0.29 | 0.15 | |
| Cell lines | Incubation time | IC50 DOX+Verapamil | IC50 T80-DOX+Verapamil | IC50 PLF127-DOX+Verapamil |
| MCF-7 | 24 h | 1.65 | 1.21 | 1.76 |
| 48 h | 1.29 | 0.67 | 0.30 | |
| 72 h | 0.29 | 0.23 | 0.19 | |
| MDA-MB-231 | 24 h | 4.3 | 0.86 | 0.55 |
| 48 h | 1.1 | 0.21 | 0.21 | |
| 72 h | 0.21 | 0.11 | 0.19 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gagliardi, A.; Cosco, D.; Udongo, B.P.; Dini, L.; Viglietto, G.; Paolino, D. Design and Characterization of Glyceryl Monooleate-Nanostructures Containing Doxorubicin Hydrochloride. Pharmaceutics 2020, 12, 1017. https://doi.org/10.3390/pharmaceutics12111017
Gagliardi A, Cosco D, Udongo BP, Dini L, Viglietto G, Paolino D. Design and Characterization of Glyceryl Monooleate-Nanostructures Containing Doxorubicin Hydrochloride. Pharmaceutics. 2020; 12(11):1017. https://doi.org/10.3390/pharmaceutics12111017
Chicago/Turabian StyleGagliardi, Agnese, Donato Cosco, Betty P. Udongo, Luciana Dini, Giuseppe Viglietto, and Donatella Paolino. 2020. "Design and Characterization of Glyceryl Monooleate-Nanostructures Containing Doxorubicin Hydrochloride" Pharmaceutics 12, no. 11: 1017. https://doi.org/10.3390/pharmaceutics12111017