Evolution from Covalent to Self-Assembled PAMAM-Based Dendrimers as Nanovectors for siRNA Delivery in Cancer by Coupled in Silico-Experimental Studies. Part II: Self-Assembled siRNA Nanocarriers
Abstract
:1. Self-Assembling PAMAM-Based Amphiphilic Dendrons: A New Paradigm for siRNA Delivery
2. siRNA Delivery by Single-Tail Self-Assembling Amphiphilic Dendrons
2.1. Design, Optimization and Chemico-Physical Characterization of Single-Tail Self-Assembling Amphiphilic Dendrons
2.2. In Silico/Experimental Interaction of Single Tail Self-Assembling Dendrons and siRNA
2.3. In Vitro siRNA Delivery Performance of Single-Tail Self-Assembling Dendrons
2.4. In Vivo siRNA Delivery Performance of Single-Tail Self-Assembling Dendron 4
3. siRNA Delivery by Double-Tail Self-Assembling Amphiphilic Dendrons
3.1. Design, Optimization and Chemico-Physical Charactrization of Double-Tail Self-Assembling Amphiphilic Dendrons
3.2. In Vitro siRNA Delivery Performance of the Double-Tail Self-Assembling Dendron AD
3.3. In Vivo siRNA Delivery Performance of the Double-Tail Self-Assembling Dendron AD
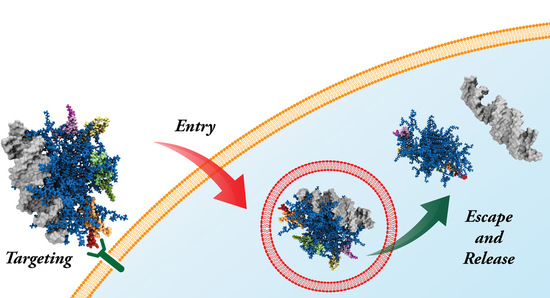
4. siRNA Delivery by the Double-Tail, Dual Targeting Self-Assembling Amphiphilic Dendron AD/E16G6RGDK
4.1. Design, Optimization and Chemico-Physical Charactrization of the Double-Tail, Dual Targeting Self-Assembling Amphiphilic Dendron AD/E16G6RGDK
4.2. In Vitro Targeted siRNA Delivery Performance of Double-Tail, Dual Targeting Self-Assembling Amphiphilic Dendron AD/E16G6RGDK
4.3. In Vivo Targeted siRNA Delivery Performance of Double-Tail, Dual Targeting Self-Assembling Amphiphilic Dendron AD/E16G6RGDK
5. Conclusions
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
Appendix A





References
- Lehn, J.-M. Toward self-organization and complex matter. Science 2002, 295, 2400–2403. [Google Scholar] [CrossRef] [PubMed]
- Whitesides, G.M.; Grzybowski, B. Self-Assembly at all scales. Science 2002, 295, 2418–2421. [Google Scholar] [CrossRef] [PubMed]
- Mattia, E.; Otto, S. Supramolecular systems chemistry. Nat. Nanotechnol. 2015, 10, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Ariga, K.; Nishikawa, M.; Mori, T.; Takeya, J.; Shrestha, L.K.; Hill, J.P. Self-assembly as a key player for materials nanoarchitectonics. Sci. Technol. Adv. Mater. 2019, 20, 51–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitty, A. Cooperativity and biological complexity. Nat. Chem. Biol. 2008, 4, 435–439. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, J.; Yu, T.; Chen, C.; Cheng, Q.; Sengupt, K.; Huang, Y.; Li, H.; Liu, C.; Wang, Y.; et al. Adaptive amphiphilic dendrimer-based nanoassemblies as robust and versatile siRNA delivery systems. Angew. Chem. Int. Ed. Engl. 2014, 53, 11822–11827. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Posocco, P.; Liu, X.; Cheng, Q.; Laurini, E.; Zhou, J.; Liu, C.; Wang, Y.; Tang, J.; Dal Col, V.; et al. Mastering dendrimer self-assembly for efficient siRNA delivery: From conceptual design to in vivo efficient gene silencing. Small 2016, 12, 3667–3676. [Google Scholar] [CrossRef]
- Dong, Y.; Yu, T.; Ding, L.; Laurini, E.; Huang, Y.; Zhang, M.; Weng, Y.; Lin, S.; Chen, P.; Marson, D.; et al. A dual targeting dendrimer-mediated siRNA delivery system for effective gene silencing in cancer therapy. J. Am. Chem. Soc. 2018, 140, 16264–16274. [Google Scholar] [CrossRef]
- Marson, D.; Laurini, E.; Aulic, S.; Fermeglia, M.; Pricl, S. Evolution from covalent to self-assembled PAMAM-based dendrimers as nanovectors for siRNA delivery in cancer by coupled in silico-experimental studies. Part I: Covalent siRNA nanocarriers. Pharmaceutics 2019, in press. [Google Scholar]
- Karatasos, K.; Posocco, P.; Laurini, E.; Pricl, S. Poly(amidoamine)-based dendrimer/siRNA complexation studied by computer simulations: Effects of pH and generation on dendrimer structure and siRNA binding. Macromol. Biosci. 2012, 12, 225–240. [Google Scholar] [CrossRef]
- Posocco, P.; Laurini, E.; Dal Col, V.; Marson, D.; Karatasos, K.; Fermeglia, M.; Pricl, S. Tell me something that I do not know. Multiscale molecular modeling of dendrimer/dendron organization and self-assembly in gene therapy. Curr. Med. Chem. 2012, 19, 5062–5087. [Google Scholar] [CrossRef] [PubMed]
- Posocco, P.; Laurini, E.; Dal Col, V.; Marson, D.; Peng, L.; Smith, D.K.; Klajnert, B.; Bryszewska, M.; Caminade, A.-M.; Majoral, J.P.; et al. Multiscale modeling of dendrimers and dendrons for drug and nucleic acid delivery. In Dendrimers in Biomedical Applications; Klajnert, B., Peng, L., Ceña, V., Eds.; RSC Publishing: Cambrige, UK, 2013; pp. 148–166. [Google Scholar]
- Pavan, G.M.; Posocco, P.; Tagliabue, A.; Maly, M.; Malek, A.; Danani, A.; Ragg, E.; Catapano, C.V.; Pricl, S. PAMAM dendrimers for siRNA delivery: Computational and experimental insights. Chem. Eur. J. 2010, 16, 7781–7795. [Google Scholar] [CrossRef] [PubMed]
- Marson, D.; Laurini, E.; Posocco, P.; Fermeglia, M.; Pricl, S. Cationic carbosilane dendrimers and oligonucleotide binding: An energetic affair. Nanoscale 2015, 7, 3876–3887. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, F.S.; Hirsch, O.; Zeisig, R.; Posocco, P.; Laurini, E.; Pricl, S.; Haag, R.; Kemmner, W.; Calderón, M. Structure–activity relationship study of dendritic polyglycerolamines for efficient siRNA transfection. RSC Adv. 2015, 5, 78760–78770. [Google Scholar] [CrossRef]
- Percec, V.; Wilson, D.A.; Leowanawat, P.; Wilson, C.J.; Huges, A.D.; Kaucher, M.S.; Hammer, D.A.; Levine, D.H.; Kim, A.J.; Bates, F.S.; et al. Self-assembly of Janus dendrimers into uniform dendrimersomes and other complex architectures. Science 2010, 328, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Rocchi, P.; So, A.; Kojima, S.; Signaevsky, M.; Beraldi, E.; Fazli, L.; Hurtado-Coll, A.; Yamanaka, K.; Gleave, M. Heat shock protein 27 increases after androgen ablation and plays a cytoprotective role in hormone-refractory prostate cancer. Cancer Res. 2004, 64, 6595–6602. [Google Scholar] [CrossRef] [PubMed]
- Behr, J.P. The proton sponge: A trick to enter cells viruses did not exploit. Chimia 1997, 51, 34–36. [Google Scholar]
- Nichols, J.W.; Bae, Y.H. EPR: Evidence and fallacy. J. Control. Release 2014, 190, 451–464. [Google Scholar] [CrossRef]
- Danhier, F. To exploit the tumor microenvironment: Since the EPR effect fails in the clinic, what is the future of nanomedicine? J. Control. Release 2016, 244, 108–121. [Google Scholar] [CrossRef]
- Desgrosellier, J.; Cheresh, D. Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 2010, 10, 9–22. [Google Scholar] [CrossRef]
- Teesalu, T.; Sugahara, K.; Kotamraju, V.; Ruoslahti, E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA 2009, 106, 16157–16162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velázquez Campoy, A.; Freire, E. ITC in the post-genomic era...? Priceless. Biophys. Chem. 2005, 115, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, K.A.; Laurini, E.; Pricl, S.; Smith, D.K. Enantiomeric and diastereomeric self-assembled multivalent nanostructures: Understanding the effects of chirality on binding to polyanionic heparin and DNA. Angew. Chem. Int. Ed. Engl. 2018, 57, 8530–8534. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, A.C.; Laurini, E.; Vieira, V.M.P.; Pricl, S.; Smith, D.K. Effect of buffer at nanoscale molecular recognition interfaces - electrostatic binding of biological polyanions. Chem. Commun. 2017, 53, 11580–11583. [Google Scholar] [CrossRef] [PubMed]
- Marson, D.; Laurini, E.; Fermeglia, M.; Smith, D.K.; Pricl, S. Mallard Blue binding to heparin, its SDS micelle-driven decomplexation, and interaction with human serum albumin: A combined experimental/modeling investigation. Fluid Phase Equilib. 2018, 470, 259–267. [Google Scholar] [CrossRef]
- Goel, H.L.; Li, J.; Kogan, S.; Languino, L.R. Integrins in prostate cancer progression. Endocr. Relat. Cancer 2008, 15, 657–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soker, S.; Takashima, S.; Miao, H.Q.; Neufeld, G.; Klagsbrun, M. Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 1998, 92, 735–745. [Google Scholar] [CrossRef]
- Available online: https://www.nature.com/subjects/self-assembly (accessed on 4 July 2019).





















| Dendron | Nagg | D | Dexp | CMC | CMCexp | ΔGm |
|---|---|---|---|---|---|---|
| 1 | 5 | 5.6 ± 0.1 | - | 415 | 398 ± 22 | −38.7 |
| 2 | 6 | 5.9 ± 0.1 | - | 127 | 116 ± 11 | −44.4 |
| 3 | 7 | 6.2 ± 0.1 | 6.6 ± 0.1 | 37.2 | 49.9 ± 3.3 | −50.5 |
| 4 | 8 | 6.4 ± 0.1 | 6.8 ± 0.1 | 11.4 | 15.6 ± 1.1 | −56.4 |
| 5 | 10 | 7.1 ± 0.2 | 7.3 ± 0.1 | 9.13 | 11.2 ± 0.6 | −57.5 |
| 6 | 13 | 8.1 ± 0.2 | 7.8 ± 0.2 | 8.50 | 8.24 ± 3.8 | −57.8 |
| Dendron | ΔGbind | Neff | ΔGbind,eff | ΔGbind,eff/Neff | C50 |
|---|---|---|---|---|---|
| 1 | −8.23 ± 0.91 | 13 ± 1 | −2.47 ± 0.32 | −0.19 ± 0.03 | - |
| 2 | −13.4 ± 1.1 | 18 ± 1 | −4.30 ± 0.45 | −0.24 ± 0.03 | 42.0 ± 2.0 |
| 3 | −17.1 ± 1.3 | 22 ± 1 | −6.00 ± 0.36 | −0.27 ± 0.02 | 16.9 ± 0.5 |
| 4 | −22.6 ± 1.2 | 31 ± 1 | −12.7 ± 0.41 | −0.41 ± 0.04 | 7.0 ± 0.3 |
| 5 | −46.6 ± 1.6 | 49 ± 1 | −28.9 ± 0.70 | −0.59 ± 0.04 | 5.3 ± 0.4 |
| 6 | −57.8 ± 2.3 | 62 ± 1 | −34.7 ± 0.62 | −0.56 ± 0.05 | 4.8 ± 0.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Laurini, E.; Marson, D.; Aulic, S.; Fermeglia, M.; Pricl, S. Evolution from Covalent to Self-Assembled PAMAM-Based Dendrimers as Nanovectors for siRNA Delivery in Cancer by Coupled in Silico-Experimental Studies. Part II: Self-Assembled siRNA Nanocarriers. Pharmaceutics 2019, 11, 324. https://doi.org/10.3390/pharmaceutics11070324
Laurini E, Marson D, Aulic S, Fermeglia M, Pricl S. Evolution from Covalent to Self-Assembled PAMAM-Based Dendrimers as Nanovectors for siRNA Delivery in Cancer by Coupled in Silico-Experimental Studies. Part II: Self-Assembled siRNA Nanocarriers. Pharmaceutics. 2019; 11(7):324. https://doi.org/10.3390/pharmaceutics11070324
Chicago/Turabian StyleLaurini, Erik, Domenico Marson, Suzana Aulic, Maurizio Fermeglia, and Sabrina Pricl. 2019. "Evolution from Covalent to Self-Assembled PAMAM-Based Dendrimers as Nanovectors for siRNA Delivery in Cancer by Coupled in Silico-Experimental Studies. Part II: Self-Assembled siRNA Nanocarriers" Pharmaceutics 11, no. 7: 324. https://doi.org/10.3390/pharmaceutics11070324