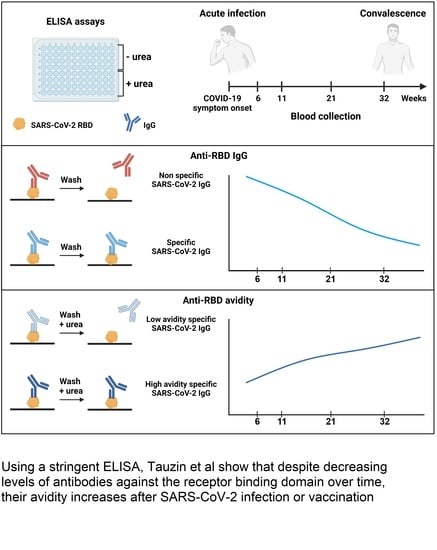
Evolution of Anti-RBD IgG Avidity following SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Plasma Samples, Primary Cells, and Antibodies
2.3. Plasmids
2.4. Cell Line, Proteins Expression and Purification
2.5. ELISA Assays
2.6. Detection of Antigen-Specific B Cells
3. Results
3.1. Cohort of COVID-19 Convalescent Donors
3.2. RBD Avidity Assay
3.3. Evolution of Anti-RBD Avidity after Resolution of Symptoms
3.4. Evolution of RBD Avidity in SARS-CoV-2 Vaccinated Individuals
3.5. RBD Avidity Correlates with B Cell Class Switch
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ding, S.; Adam, D.; Beaudoin-Bussières, G.; Tauzin, A.; Gong, S.Y.; Gasser, R.; Laumaea, A.; Anand, S.P.; Privé, A.; Bourassa, C.; et al. SARS-CoV-2 Spike Expression at the Surface of Infected Primary Human Airway Epithelial Cells. Viruses 2021, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Characterization of Spike Glycoprotein of SARS-CoV-2 on Virus Entry and Its Immune Cross-Reactivity with SARS-CoV. Nat. Commun. 2020, 11, 1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural Basis of Receptor Recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human Neutralizing Antibodies Elicited by SARS-CoV-2 Infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Min, L.; Sun, Q. Antibodies and Vaccines Target RBD of SARS-CoV-2. Front. Mol. Biosci. 2021, 8, 671633. [Google Scholar] [CrossRef]
- Anand, S.P.; Prévost, J.; Nayrac, M.; Beaudoin-Bussières, G.; Benlarbi, M.; Gasser, R.; Brassard, N.; Laumaea, A.; Gong, S.Y.; Bourassa, C.; et al. Longitudinal Analysis of Humoral Immunity against SARS-CoV-2 Spike in Convalescent Individuals up to Eight Months Post-Symptom Onset. Cell Rep. Med. 2021, 2, 100290. [Google Scholar] [CrossRef]
- Beaudoin-Bussières, G.; Laumaea, A.; Anand, S.P.; Prévost, J.; Gasser, R.; Goyette, G.; Medjahed, H.; Perreault, J.; Tremblay, T.; Lewin, A.; et al. Decline of Humoral Responses against SARS-CoV-2 Spike in Convalescent Individuals. MBio 2020, 11, e02590-20. [Google Scholar] [CrossRef]
- Prévost, J.; Gasser, R.; Beaudoin-Bussières, G.; Richard, J.; Duerr, R.; Laumaea, A.; Anand, S.P.; Goyette, G.; Benlarbi, M.; Ding, S.; et al. Cross-Sectional Evaluation of Humoral Responses against SARS-CoV-2 Spike. Cell Rep. Med. 2020, 1, 100126. [Google Scholar] [CrossRef]
- Goel, R.R.; Painter, M.M.; Apostolidis, S.A.; Mathew, D.; Meng, W.; Rosenfeld, A.M.; Lundgreen, K.A.; Reynaldi, A.; Khoury, D.S.; Pattekar, A.; et al. MRNA Vaccines Induce Durable Immune Memory to SARS-CoV-2 and Variants of Concern. Science 2021, 374, eabm0829. [Google Scholar] [CrossRef] [PubMed]
- Tauzin, A.; Gong, S.Y.; Beaudoin-Bussières, G.; Vézina, D.; Gasser, R.; Nault, L.; Marchitto, L.; Benlarbi, M.; Chatterjee, D.; Nayrac, M.; et al. Strong Humoral Immune Responses against SARS-CoV-2 Spike after BNT162b2 MRNA Vaccination with a 16-Week Interval between Doses. Cell Host Microbe 2021, 30, 97–109.e5. [Google Scholar] [CrossRef] [PubMed]
- Xiang, T.; Liang, B.; Fang, Y.; Lu, S.; Li, S.; Wang, H.; Li, H.; Yang, X.; Shen, S.; Zhu, B.; et al. Declining Levels of Neutralizing Antibodies Against SARS-CoV-2 in Convalescent COVID-19 Patients One Year Post Symptom Onset. Front. Immunol. 2021, 12, 2327. [Google Scholar] [CrossRef] [PubMed]
- Vicenti, I.; Basso, M.; Gatti, F.; Scaggiante, R.; Boccuto, A.; Zago, D.; Modolo, E.; Dragoni, F.; Parisi, S.G.; Zazzi, M. Faster Decay of Neutralizing Antibodies in Never Infected than Previously Infected Healthcare Workers Three Months after the Second BNT162b2 MRNA COVID-19 Vaccine Dose. Int. J. Infect. Dis. 2021, 112, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Bayart, J.-L.; Douxfils, J.; Gillot, C.; David, C.; Mullier, F.; Elsen, M.; Eucher, C.; Van Eeckhoudt, S.; Roy, T.; Gerin, V.; et al. Waning of IgG, Total and Neutralizing Antibodies 6 Months Post-Vaccination with BNT162b2 in Healthcare Workers. Vaccines 2021, 9, 1092. [Google Scholar] [CrossRef] [PubMed]
- Muecksch, F.; Weisblum, Y.; Barnes, C.O.; Schmidt, F.; Schaefer-Babajew, D.; Wang, Z.; C Lorenzi, J.C.; Flyak, A.I.; DeLaitsch, A.T.; Huey-Tubman, K.E.; et al. Affinity Maturation of SARS-CoV-2 Neutralizing Antibodies Confers Potency, Breadth, and Resilience to Viral Escape Mutations. Immunity 2021, 54, 1853–1868.e7. [Google Scholar] [CrossRef]
- Cho, A.; Muecksch, F.; Schaefer-Babajew, D.; Wang, Z.; Finkin, S.; Gaebler, C.; Ramos, V.; Cipolla, M.; Mendoza, P.; Agudelo, M.; et al. Anti-SARS-CoV-2 Receptor-Binding Domain Antibody Evolution after MRNA Vaccination. Nature 2021, 600, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Muecksch, F.; Schaefer-Babajew, D.; Finkin, S.; Viant, C.; Gaebler, C.; Hoffmann, H.-H.; Barnes, C.O.; Cipolla, M.; Ramos, V.; et al. Naturally Enhanced Neutralizing Breadth against SARS-CoV-2 One Year after Infection. Nature 2021, 595, 426–431. [Google Scholar] [CrossRef]
- Gaebler, C.; Wang, Z.; Lorenzi, J.C.C.; Muecksch, F.; Finkin, S.; Tokuyama, M.; Cho, A.; Jankovic, M.; Schaefer-Babajew, D.; Oliveira, T.Y.; et al. Evolution of Antibody Immunity to SARS-CoV-2. Nature 2021, 591, 639–644. [Google Scholar] [CrossRef]
- Bachmann, M.F.; Kalinke, U.; Althage, A.; Freer, G.; Burkhart, C.; Roost, H.-P.; Aguet, M.; Hengartner, H.; Zinkernagel, R.M. The Role of Antibody Concentration and Avidity in Antiviral Protection. Science 1997, 276, 2024–2027. [Google Scholar] [CrossRef]
- Victora, G.D.; Nussenzweig, M.C. Germinal Centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Björkman, C.; Näslund, K.; Stenlund, S.; Maley, S.W.; Buxton, D.; Uggla, A. An IgG Avidity ELISA to Discriminate between Recent and Chronic Neospora Caninum Infection. J. Vet. Diagn. Investig. 1999, 11, 41–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fialová, L.; Petráčková, M.; Kuchař, O. Comparison of Different Enzyme-Linked Immunosorbent Assay Methods for Avidity Determination of Antiphospholipid Antibodies. J. Clin. Lab. Anal. 2017, 31, e22121. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Lei, Y.; Lu, X.; Wang, G.; Du, Q.; Guo, X.; Xing, Y.; Zhang, G.; Wang, D. Urea-Mediated Dissociation Alleviate the False-Positive Treponema Pallidum-Specific Antibodies Detected by ELISA. PLoS ONE 2019, 14, e0212893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perreault, J.; Tremblay, T.; Fournier, M.-J.; Drouin, M.; Beaudoin-Bussières, G.; Prévost, J.; Lewin, A.; Bégin, P.; Finzi, A.; Bazin, R. Waning of SARS-CoV-2 RBD Antibodies in Longitudinal Convalescent Plasma Samples within 4 Months after Symptom Onset. Blood 2020, 136, 2588–2591. [Google Scholar] [CrossRef] [PubMed]
- MacLennan, I.C.M. Germinal Centers. Annu. Rev. Immunol. 1994, 12, 117–139. [Google Scholar] [CrossRef]
- De Silva, N.S.; Klein, U. Dynamics of B Cells in Germinal Centres. Nat. Rev. Immunol. 2015, 15, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Ohta, H.; Takemori, T. Fas Is Required for Clonal Selection in Germinal Centers and the Subsequent Establishment of the Memory B Cell Repertoire. Immunity 2001, 14, 181–192. [Google Scholar] [CrossRef]
- Gatto, D.; Martin, S.W.; Bessa, J.; Pellicioli, E.; Saudan, P.; Hinton, H.J.; Bachmann, M.F. Regulation of Memory Antibody Levels: The Role of Persisting Antigen versus Plasma Cell Life Span. J. Immunol. 2007, 178, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Garcia-Ibanez, L.; Toellner, K.-M. Regulation of Germinal Center B-Cell Differentiation. Immunol. Rev. 2016, 270, 8–19. [Google Scholar] [CrossRef]
- Young, C.; Brink, R. The Unique Biology of Germinal Center B Cells. Immunity 2021, 54, 1652–1664. [Google Scholar] [CrossRef] [PubMed]
- Tauzin, A.; Nayrac, M.; Benlarbi, M.; Gong, S.Y.; Gasser, R.; Beaudoin-Bussières, G.; Brassard, N.; Laumaea, A.; Vézina, D.; Prévost, J.; et al. A Single Dose of the SARS-CoV-2 Vaccine BNT162b2 Elicits Fc-Mediated Antibody Effector Functions and T Cell Responses. Cell Host Microbe 2021, 29, 1137–1150. [Google Scholar] [CrossRef] [PubMed]
- Beaudoin-Bussières, G.; Chen, Y.; Ullah, I.; Prévost, J.; Tolbert, W.D.; Symmes, K.; Ding, S.; Benlarbi, M.; Gong, S.Y.; Tauzin, A.; et al. A Fc-Enhanced NTD-Binding Non-Neutralizing Antibody Delays Virus Spread and Synergizes with a NAb to Protect Mice from Lethal SARS-CoV-2 Infection. Cell Rep. 2022, 38, 110368. [Google Scholar] [CrossRef] [PubMed]
- Adeniji, O.S.; Giron, L.B.; Purwar, M.; Zilberstein, N.F.; Kulkarni, A.J.; Shaikh, M.W.; Balk, R.A.; Moy, J.N.; Forsyth, C.B.; Liu, Q.; et al. COVID-19 Severity Is Associated with Differential Antibody Fc-Mediated Innate Immune Functions. MBio 2021, 12, e00281-21. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, D.; Tauzin, A.; Marchitto, L.; Gong, S.Y.; Boutin, M.; Bourassa, C.; Beaudoin-Bussières, G.; Bo, Y.; Ding, S.; Laumaea, A.; et al. SARS-CoV-2 Omicron Spike Recognition by Plasma from Individuals Receiving BNT162b2 MRNA Vaccination with a 16-Week Interval between Doses. Cell Rep. 2022, 110429. [Google Scholar] [CrossRef] [PubMed]




| Group | n | Days PSO (Median; Day Range) | Age (Median; Age Range) | Male (n) | Female (n) |
|---|---|---|---|---|---|
| 6 weeks | 29 | 45 (16–95) | 48 (21–65) | 15 | 14 |
| 11 weeks | 25 | 77 (48–127) | 48 (21–65) | 14 | 11 |
| 21 weeks | 26 | 146 (116–171) | 49 (21–65) | 14 | 12 |
| 32 weeks | 24 | 226 (201–275) | 51 (21–65) | 15 | 9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tauzin, A.; Gendron-Lepage, G.; Nayrac, M.; Anand, S.P.; Bourassa, C.; Medjahed, H.; Goyette, G.; Dubé, M.; Bazin, R.; Kaufmann, D.E.; et al. Evolution of Anti-RBD IgG Avidity following SARS-CoV-2 Infection. Viruses 2022, 14, 532. https://doi.org/10.3390/v14030532
Tauzin A, Gendron-Lepage G, Nayrac M, Anand SP, Bourassa C, Medjahed H, Goyette G, Dubé M, Bazin R, Kaufmann DE, et al. Evolution of Anti-RBD IgG Avidity following SARS-CoV-2 Infection. Viruses. 2022; 14(3):532. https://doi.org/10.3390/v14030532
Chicago/Turabian StyleTauzin, Alexandra, Gabrielle Gendron-Lepage, Manon Nayrac, Sai Priya Anand, Catherine Bourassa, Halima Medjahed, Guillaume Goyette, Mathieu Dubé, Renée Bazin, Daniel E. Kaufmann, and et al. 2022. "Evolution of Anti-RBD IgG Avidity following SARS-CoV-2 Infection" Viruses 14, no. 3: 532. https://doi.org/10.3390/v14030532