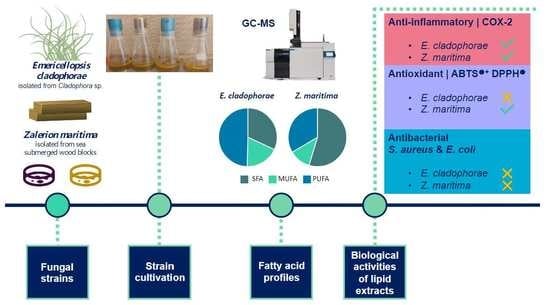
Evaluation of Lipid Extracts from the Marine Fungi Emericellopsis cladophorae and Zalerion maritima as a Source of Anti-Inflammatory, Antioxidant and Antibacterial Compounds
Abstract
:1. Introduction
2. Results
2.1. Fatty Acid Profiles
2.2. Anti-Inflammatory Activity
2.3. Antioxidant Activity
2.4. Antibacterial Activity
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Fungal Strains
4.3. Strain Cultivation
4.4. Lipid Extraction
4.5. Phospholipid Quantification
4.6. Fatty Acid Analysis through Gas Chromatography—Mass Spectrometry (GC-MS)
4.6.1. Transesterification
4.6.2. Gas Chromatography—Mass Spectrometry (GC-MS)
4.6.3. Identification and Integration
4.7. Determination of Anti-Inflammatory Activity of Lipid Extracts
4.8. Determination of Antioxidant Activity of Lipid Extracts
4.8.1. DPPH Radical Scavenging Activity Assay
4.8.2. ABTS Radical Scavenging Activity Assay
4.9. Determination of Antibacterial Activity of Lipid Extracts
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Debbab, A.; Aly, A.H.; Lin, W.H.; Proksch, P. Bioactive Compounds from Marine Bacteria and Fungi. Microb. Biotechnol. 2010, 3, 544–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deshmukh, S.K.; Prakash, V.; Ranjan, N. Marine Fungi: A Source of Potential Anticancer Compounds. Front. Microbiol. 2018, 8, 2536. [Google Scholar] [CrossRef] [PubMed]
- Hasan, S.; Ansari, M.; Ahmad, A.; Mishra, M. Major Bioactive Metabolites from Marine Fungi: A Review. Bioinformation 2015, 11, 176–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, E.; Dias, M.; Lopes, D.; Almeida, A.; Domingues, M.D.R.; Rey, F. Antimicrobial Lipids from Plants and Marine Organisms: An Overview of the Current State-of-the-Art and Future Prospects. Antibiotics 2020, 9, 441. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2022, 39, 1122. [Google Scholar] [CrossRef]
- Athenaki, M.; Gardeli, A.; Diamantopoulou, P.; Tchakouteu, S.; Sarris, D.; Philippoussis, A.; Papanikolaou, S. Lipids from Yeasts and Fungi: Physiology, Production and Analytical Considerations. J. Appl. Microbiol. 2017, 124, 336–367. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, M.; Aleixo, A.; Vicente, F.; Esteves, A.; Alves, A. Three New Species of Neocamarosporium Isolated from Saline Environments: N. aestuarinum Sp. Nov., N. endophyticum Sp. Nov. and N. halimiones Sp. Nov. Mycosphere 2019, 10, 608–621. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Vicente, T.F.L.; Esteves, A.C.; Alves, A. Novel Halotolerant Species of Emericellopsis and Parasarocladium Associated with Macroalgae in an Estuarine Environment. Mycologia 2020, 112, 154–171. [Google Scholar] [CrossRef]
- Marchese, P.; Garzoli, L.; Young, R.; Allcock, L.; Barry, F.; Tuohy, M.; Murphy, M. Fungi Populate Deep-sea Coral Gardens as Well as Marine Sediments in the Irish Atlantic Ocean. Environ. Microbiol. 2021, 23, 4168–4184. [Google Scholar] [CrossRef]
- Abdel-Wahab, M.A.; Jones, E.B.G.; Bahkali, A.H.A.; El-Gorban, A.M. Marine Fungi from Red Sea Mangroves in Saudi Arabia with Fulvocentrum rubrum sp. nov. (Torpedosporales, Ascomycota). Nova_Hedwig. 2019, 108, 365–377. [Google Scholar] [CrossRef]
- Gladfelter, A.S.; James, T.Y.; Amend, A.S. Marine Fungi. Curr. Biol. 2019, 29, R191–R195. [Google Scholar] [CrossRef]
- Amend, A.; Burgaud, G.; Cunliffe, M.; Edgcomb, V.P.; Ettinger, C.L.; Gutiérrez, M.H.; Heitman, J.; Hom, E.F.Y.; Ianiri, G.; Jones, A.C.; et al. Fungi in the Marine Environment: Open Questions and Unsolved Problems. mBio 2019, 10, e01189-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Strat, Y.; Ruiz, N.; Fleurence, J.; Pouchus, Y.-F.; Déléris, P.; Dumay, J. Marine Fungal Abilities to Enzymatically Degrade Algal Polysaccharides, Proteins and Lipids: A Review. J. Appl. Phycol. 2022, 34, 1131–1162. [Google Scholar] [CrossRef]
- Savidov, N.; Gloriozova, T.A.; Poroikov, V.V.; Dembitsky, V.M. Highly Oxygenated Isoprenoid Lipids Derived from Fungi and Fungal Endophytes: Origin and Biological Activities. Steroids 2018, 140, 114–124. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Chen, J.; Hu, G.; Yu, J.; Zhu, X.; Lin, Y.; Chen, S.; Yuan, J. Statistical Research on the Bioactivity of New Marine Natural Products Discovered during the 28 Years from 1985 to 2012. Mar. Drugs 2015, 13, 202–221. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.; Rämä, T.; Oosterhuis, R.; Walker, A.; Pang, K.-L. The Neglected Marine Fungi, Sensu Stricto, and Their Isolation for Natural Products’ Discovery. Mar. Drugs 2019, 17, 42. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Yi, M.; Ding, L.; He, S. A Review of Anti-Inflammatory Compounds from Marine Fungi, 2000–2018. Mar. Drugs 2019, 17, 636. [Google Scholar] [CrossRef] [Green Version]
- Wiese, J.; Imhoff, J.F. Marine Bacteria and Fungi as Promising Source for New Antibiotics. Drug. Dev. Res. 2019, 80, 24–27. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, M.F.M.; Paço, A.; Escada, L.F.; Albuquerque, M.S.F.; Pinto, C.A.; Saraiva, J.A.; Duarte, A.S.; Rocha-Santos, T.A.P.; Esteves, A.C.; Alves, A. Unveiling Biological Activities of Marine Fungi: The Effect of Sea Salt. Appl. Sci. 2021, 11, 6008. [Google Scholar] [CrossRef]
- Ferreira, J.A.; Lennartsson, P.R.; Edebo, L.; Taherzadeh, M.J. Zygomycetes-Based Biorefinery: Present Status and Future Prospects. Bioresour. Technol. 2013, 135, 523–532. [Google Scholar] [CrossRef] [Green Version]
- Tsoupras, A.; Kouvelis, V.N.; Pappas, K.M.; Demopoulos, C.A.; Typas, M.A. Anti-Inflammatory and Anti-Thrombotic Properties of Lipid Bioactives from the Entomopathogenic Fungus Beauveria bassiana. Prostaglandins Other Lipid Mediat. 2022, 158, 106606. [Google Scholar] [CrossRef] [PubMed]
- Codoñer-Franch, P.; Valls-Bellés, V.; Arilla-Codoñer, A.; Alonso-Iglesias, E. Oxidant Mechanisms in Childhood Obesity: The Link between Inflammation and Oxidative Stress. Transl. Res. 2011, 158, 369–384. [Google Scholar] [CrossRef]
- Steven, S.; Frenis, K.; Oelze, M.; Kalinovic, S.; Kuntic, M.; Bayo Jimenez, M.T.; Vujacic-Mirski, K.; Helmstädter, J.; Kröller-Schön, S.; Münzel, T.; et al. Vascular Inflammation and Oxidative Stress: Major Triggers for Cardiovascular Disease. Oxidative Med. Cell. Longev. 2019, 2019, 7092151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rawdin, B.J.; Mellon, S.H.; Dhabhar, F.S.; Epel, E.S.; Puterman, E.; Su, Y.; Burke, H.M.; Reus, V.I.; Rosser, R.; Hamilton, S.P.; et al. Dysregulated Relationship of Inflammation and Oxidative Stress in Major Depression. Brain Behav. Immun. 2013, 31, 143–152. [Google Scholar] [CrossRef] [Green Version]
- Krumova, K.; Cosa, G. Chapter 1. Overview of Reactive Oxygen Species. In Comprehensive Series in Photochemical & Photobiological Sciences; Nonell, S., Flors, C., Eds.; Royal Society of Chemistry: Cambridge, UK, 2016; Volume 1, pp. 1–21. ISBN 978-1-78262-038-9. [Google Scholar]
- Prasad, S.; Gupta, S.C.; Tyagi, A.K. Reactive Oxygen Species (ROS) and Cancer: Role of Antioxidative Nutraceuticals. Cancer Lett. 2017, 387, 95–105. [Google Scholar] [CrossRef]
- Giorgi, C.; Marchi, S.; Simoes, I.C.M.; Ren, Z.; Morciano, G.; Perrone, M.; Patalas-Krawczyk, P.; Borchard, S.; Jędrak, P.; Pierzynowska, K.; et al. Mitochondria and Reactive Oxygen Species in Aging and Age-Related Diseases. Int. Rev. Cell Mol. Biol. 2018, 340, 209–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.; Lee, Y.S.; Jung, S.H.; Kang, S.S.; Shin, K.H. Anti-Oxidant Activities of Fucosterol from the Marine Algae Pelvetia siliquosa. Arch. Pharm. Res 2003, 26, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Terme, N.; Boulho, R.; Kucma, J.-P.; Bourgougnon, N.; Bedoux, G. Radical Scavenging Activity of Lipids from Seaweeds Isolated by Solid-Liquid Extraction and Supercritical Fluids. OCL 2018, 25, D505. [Google Scholar] [CrossRef]
- Ha, A.W.; Na, S.J.; Kim, W.K. Antioxidant Effects of Fucoxanthin Rich Powder in Rats Fed with High Fat Diet. Nutr. Res. Pract. 2013, 7, 475. [Google Scholar] [CrossRef] [Green Version]
- Lordan, S.; Ross, R.P.; Stanton, C. Marine Bioactives as Functional Food Ingredients: Potential to Reduce the Incidence of Chronic Diseases. Mar. Drugs 2011, 9, 1056–1100. [Google Scholar] [CrossRef] [Green Version]
- Pallela, R. Antioxidants from Marine Organisms and Skin Care. In Systems Biology of Free Radicals and Antioxidants; Laher, I., Ed.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3771–3783. ISBN 978-3-642-30017-2. [Google Scholar]
- Couttolenc, A.; Medina, M.E.; Trigos, Á.; Espinoza, C. Antioxidant Capacity of Fungi Associated with Corals and Sponges of the Reef System of Veracruz, Mexico. Electron. J. Biotechnol. 2022, 55, 40–46. [Google Scholar] [CrossRef]
- Sun, H.-H.; Mao, W.-J.; Chen, Y.; Guo, S.-D.; Li, H.-Y.; Qi, X.-H.; Chen, Y.-L.; Xu, J. Isolation, Chemical Characteristics and Antioxidant Properties of the Polysaccharides from Marine Fungus Penicillium sp. F23-2. Carbohydr. Polym. 2009, 78, 117–124. [Google Scholar] [CrossRef]
- Paço, A.; Duarte, K.; da Costa, J.P.; Santos, P.S.M.; Pereira, R.; Pereira, M.E.; Freitas, A.C.; Duarte, A.C.; Rocha-Santos, T.A.P. Biodegradation of Polyethylene Microplastics by the Marine Fungus Zalerion maritimum. Sci. Total Environ. 2017, 586, 10–15. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Hilário, S.; Van de Peer, Y.; Esteves, A.C.; Alves, A. Genomic and Metabolomic Analyses of the Marine Fungus Emericellopsis cladophorae: Insights into Saltwater Adaptability Mechanisms and Its Biosynthetic Potential. J. Fungi 2022, 8, 31. [Google Scholar] [CrossRef]
- Thomas, S.; Lengger, S.K.; Bird, K.E.; Allen, R.; Cunliffe, M. Macromolecular Composition and Substrate Range of Three Marine Fungi across Major Cell Types. FEMS Microbes 2022, 3, xtab019. [Google Scholar] [CrossRef]
- Subramaniam, R.; Dufreche, S.; Zappi, M.; Bajpai, R. Microbial Lipids from Renewable Resources: Production and Characterization. J. Ind. Microbiol. Biotechnol. 2010, 37, 1271–1287. [Google Scholar] [CrossRef] [PubMed]
- Fakas, S.; Makri, A.; Mavromati, M.; Tselepi, M.; Aggelis, G. Fatty Acid Composition in Lipid Fractions Lengthwise the Mycelium of Mortierella isabellina and Lipid Production by Solid State Fermentation. Bioresour. Technol. 2009, 100, 6118–6120. [Google Scholar] [CrossRef]
- Gutiérrez, M.; Vera, J.; Srain, B.; Quiñones, R.; Wörmer, L.; Hinrichs, K.; Pantoja-Gutiérrez, S. Biochemical Fingerprints of Marine Fungi: Implications for Trophic and Biogeochemical Studies. Aquat. Microb. Ecol. 2020, 84, 75–90. [Google Scholar] [CrossRef] [Green Version]
- Cooney, J.J.; Doolittle, M.M.; Grahl-Nielsen, O.; Haaland, I.M.; Kirk, P.W. Comparison of Fatty Acids of Marine Fungi Using Multivariate Statistical Analysis. J. Ind. Microbiol. 1993, 12, 373–378. [Google Scholar] [CrossRef]
- Kelly, J.; Scheibling, R. Fatty Acids as Dietary Tracers in Benthic Food Webs. Mar. Ecol. Prog. Ser. 2012, 446, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Pandohee, J. Alpha-Linolenic Acid. In Nutraceuticals and Health Care; Elsevier: Amsterdam, The Netherlands, 2022; pp. 279–288. ISBN 978-0-323-89779-2. [Google Scholar]
- Yuan, Q.; Xie, F.; Huang, W.; Hu, M.; Yan, Q.; Chen, Z.; Zheng, Y.; Liu, L. The Review of Alpha-linolenic Acid: Sources, Metabolism, and Pharmacology. Phytother. Res. 2022, 36, 164–188. [Google Scholar] [CrossRef]
- Simard, M.; Tremblay, A.; Morin, S.; Martin, C.; Julien, P.; Fradette, J.; Flamand, N.; Pouliot, R. α-Linolenic Acid and Linoleic Acid Modulate the Lipidome and the Skin Barrier of a Tissue-Engineered Skin Model. Acta Biomater. 2022, 140, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Alexanian, A.; Sorokin, A. Cyclooxygenase 2: Protein-Protein Interactions and Posttranslational Modifications. Physiol Genom. 2017, 49, 667–681. [Google Scholar] [CrossRef]
- Conde, T.A.; Zabetakis, I.; Tsoupras, A.; Medina, I.; Costa, M.; Silva, J.; Neves, B.; Domingues, P.; Domingues, M.R. Microalgal Lipid Extracts Have Potential to Modulate the Inflammatory Response: A Critical Review. IJMS 2021, 22, 9825. [Google Scholar] [CrossRef] [PubMed]
- Pauls, S.D.; Rodway, L.A.; Winter, T.; Taylor, C.G.; Zahradka, P.; Aukema, H.M. Anti-Inflammatory Effects of α-Linolenic Acid in M1-like Macrophages Are Associated with Enhanced Production of Oxylipins from α-Linolenic and Linoleic Acid. J. Nutr. Biochem. 2018, 57, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Caligiuri, S.P.B.; Aukema, H.M.; Ravandi, A.; Pierce, G.N. Elevated Levels of Pro-Inflammatory Oxylipins in Older Subjects Are Normalized by Flaxseed Consumption. Exp. Gerontol. 2014, 59, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chung, S.H. Anti-Inflammatory Effect of α-Linolenic Acid and Its Mode of Action through the Inhibition of Nitric Oxide Production and Inducible Nitric Oxide Synthase Gene Expression via NF-ΚB and Mitogen-Activated Protein Kinase Pathways. J. Agric. Food Chem. 2007, 55, 5073–5080. [Google Scholar] [CrossRef]
- Hameed, A.; Hussain, S.A.; Yang, J.; Ijaz, M.U.; Liu, Q.; Suleria, H.A.R.; Song, Y. Antioxidants Potential of the Filamentous Fungi (Mucor circinelloides). Nutrients 2017, 9, 1101. [Google Scholar] [CrossRef] [Green Version]
- Smith, H.; Doyle, S.; Murphy, R. Filamentous Fungi as a Source of Natural Antioxidants. Food Chem. 2015, 185, 389–397. [Google Scholar] [CrossRef]
- Papanikolaou, S.; Galiotou-Panayotou, M.; Chevalot, I.; Komaitis, M.; Marc, I.; Aggelis, G. Influence of Glucose and Saturated Free-Fatty Acid Mixtures on Citric Acid and Lipid Production by Yarrowia lipolytica. Curr. Microbiol. 2006, 52, 134–142. [Google Scholar] [CrossRef]
- Oppedisano, F.; Macrì, R.; Gliozzi, M.; Musolino, V.; Carresi, C.; Maiuolo, J.; Bosco, F.; Nucera, S.; Caterina Zito, M.; Guarnieri, L.; et al. The Anti-Inflammatory and Antioxidant Properties of n-3 PUFAs: Their Role in Cardiovascular Protection. Biomedicines 2020, 8, 306. [Google Scholar] [CrossRef] [PubMed]
- Conde, T.A.; Neves, B.F.; Couto, D.; Melo, T.; Neves, B.; Costa, M.; Silva, J.; Domingues, P.; Domingues, M.R. Microalgae as Sustainable Bio-Factories of Healthy Lipids: Evaluating Fatty Acid Content and Antioxidant Activity. Mar. Drugs 2021, 19, 357. [Google Scholar] [CrossRef]
- Bartolomeu, M.; Vieira, C.; Dias, M.; Conde, T.; Couto, D.; Lopes, D.; Neves, B.; Melo, T.; Rey, F.; Alves, E.; et al. Bioprospecting Antibiotic Properties in Photodynamic Therapy of Lipids from Codium tomemtosum and Chlorella vulgaris. Biochimie 2022, 203, 32–39. [Google Scholar] [CrossRef]
- Kuvarina, A.E.; Gavryushina, I.A.; Kulko, A.B.; Ivanov, I.A.; Rogozhin, E.A.; Georgieva, M.L.; Sadykova, V.S. The Emericellipsins A–E from an Alkalophilic Fungus Emericellopsis alkalina Show Potent Activity against Multidrug-Resistant Pathogenic Fungi. JoF 2021, 7, 153. [Google Scholar] [CrossRef]
- Gonçalves, M.F.M.; Abreu, A.C.; Hilário, S.; Alves, A. Diversity of Marine Fungi Associated with Wood Baits in the Estuary Ria de Aveiro, with Descriptions of Paralulworthia halima, comb. nov., Remispora submersa, sp. nov., and Zalerion pseudomaritima, sp. nov. Mycologia 2021, 113, 664–683. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A Rapid Method of Total Lipid Extraction and Purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Bartlett, M.E.; Lewis, D.H. Spectrophotometric Determination of Phosphate Esters in the Presence and Absence of Orthophosphate. Anal. Biochem. 1970, 36, 159–167. [Google Scholar] [CrossRef] [PubMed]
- Magalhães, L.M.; Segundo, M.A.; Reis, S.; Lima, J.L.F.C. Automatic Method for Determination of Total Antioxidant Capacity Using 2,2-Diphenyl-1-Picrylhydrazyl Assay. Anal. Chim. Acta 2006, 558, 310–318. [Google Scholar] [CrossRef]
- Rey, F.; Cartaxana, P.; Melo, T.; Calado, R.; Pereira, R.; Abreu, H.; Domingues, P.; Cruz, S.; Domingues, R.M. Domesticated Populations of Codium tomentosum Display Lipid Extracts with Lower Seasonal Shifts than Conspecifics from the Wild—Relevance for Biotechnological Applications of This Green Seaweed. Mar. Drugs 2020, 18, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magalhães, L.M.; Barreiros, L.; Maia, M.A.; Reis, S.; Segundo, M.A. Rapid Assessment of Endpoint Antioxidant Capacity of Red Wines through Microchemical Methods Using a Kinetic Matching Approach. Talanta 2012, 97, 473–483. [Google Scholar] [CrossRef] [PubMed]
- Ozgen, M.; Reese, R.N.; Tulio, A.Z.; Scheerens, J.C.; Miller, A.R. Modified 2,2-Azino-Bis-3-Ethylbenzothiazoline-6-Sulfonic Acid (ABTS) Method to Measure Antioxidant Capacity of Selected Small Fruits and Comparison to Ferric Reducing Antioxidant Power (FRAP) and 2,2′-Diphenyl-1- Picrylhydrazyl (DPPH) Methods. J. Agric. Food Chem. 2006, 54, 1151. [Google Scholar] [CrossRef]


| Fatty Acid | Emericellopsis cladophorae | Zalerion maritima |
|---|---|---|
| 14:0 | 0.16 ± 0.01 | 0.43 ± 0.06 |
| 15:0 | 0.08 ± 0.01 | 0.09 ± 0.01 |
| 16:0 | 15.97 ± 0.73 | 27.64 ± 2.72 |
| 16:1 | 0.07 ± 0.03 | — |
| 16:1 n-7 | 0.40 ± 0.04 | 0.19 ± 0.09 |
| 16:2 n-4 | 0.08 ± 0.02 | — |
| 17:0 | 0.07 ± 0.01 | 0.12 ± 0.02 |
| 18:0 | 13.51 ± 5.55 | 26.43 ± 9.34 |
| 18:1 n-9 | 18.35 ± 0.59 | 11.07 ± 3.20 |
| 18:1 | 0.17 ± 0.02 | 0.35 ± 0.10 |
| 18:2 n-6 | 45.38 ± 4.79 | 24.51 ± 7.32 |
| 18:3 n-3 | 4.22 ± 0.69 | 7.95 ± 1.47 |
| 20:0 | 0.29 ± 0.11 | 0.33 ± 0.05 |
| 20-methyl-heneicosanoate (iso) | — | 0.30 ± 0.09 |
| 20:1 | — | 0.17 ± 0.10 |
| 20:2 n-6 | 0.17 ± 0.04 | 0.12 ± 0.07 |
| 22:0 | 0.61 ± 0.04 | — |
| 24:0 | 0.48 ± 0.07 | 0.33 ± 0.10 |
| SFA | 31.16 ± 5.10 | 55.37 ± 11.75 |
| MUFA | 18.99 ± 0.58 | 11.78 ± 3.40 |
| PUFA | 49.85 ± 5.45 | 32.55 ± 8.36 |
| IC20 µg mL−1 | TE µmol Trolox g−1 Lipid Extract | |
|---|---|---|
| DPPH• | 116.63 ± 6.21 | 92.13 ± 4.82 |
| ABTS•+ | 101.30 ± 14.37 | 106.58 ± 14.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abraúl, M.; Alves, A.; Hilário, S.; Melo, T.; Conde, T.; Domingues, M.R.; Rey, F. Evaluation of Lipid Extracts from the Marine Fungi Emericellopsis cladophorae and Zalerion maritima as a Source of Anti-Inflammatory, Antioxidant and Antibacterial Compounds. Mar. Drugs 2023, 21, 199. https://doi.org/10.3390/md21040199
Abraúl M, Alves A, Hilário S, Melo T, Conde T, Domingues MR, Rey F. Evaluation of Lipid Extracts from the Marine Fungi Emericellopsis cladophorae and Zalerion maritima as a Source of Anti-Inflammatory, Antioxidant and Antibacterial Compounds. Marine Drugs. 2023; 21(4):199. https://doi.org/10.3390/md21040199
Chicago/Turabian StyleAbraúl, Mariana, Artur Alves, Sandra Hilário, Tânia Melo, Tiago Conde, Maria Rosário Domingues, and Felisa Rey. 2023. "Evaluation of Lipid Extracts from the Marine Fungi Emericellopsis cladophorae and Zalerion maritima as a Source of Anti-Inflammatory, Antioxidant and Antibacterial Compounds" Marine Drugs 21, no. 4: 199. https://doi.org/10.3390/md21040199