Targeted Isolation of Antibiotic Brominated Alkaloids from the Marine Sponge Pseudoceratina durissima Using Virtual Screening and Molecular Networking
Abstract
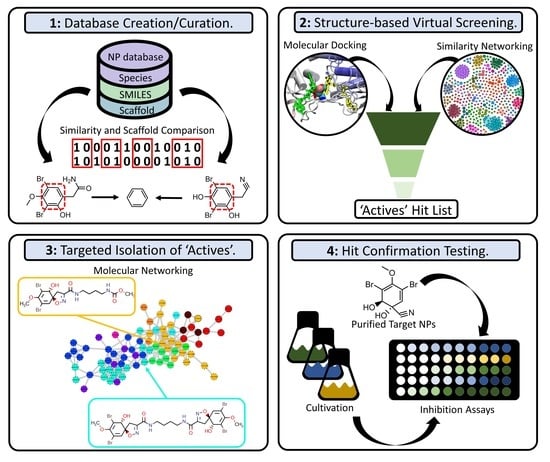
:1. Introduction
2. Results and Discussion
2.1. Data Set Curation and Treatment
2.2. Docking-Based Virtual Screening
2.3. Molecular Networking and Compound Isolation
2.4. Bioactivity Testing of Extracts and Compounds
2.4.1. Antimicrobial Assay Results
2.4.2. Anthelmintic Assay Results
2.4.3. Zebrafish Assay Results
2.5. Molecular Docking of (+)-Aeroplysinin-1
2.6. Limitations and Perspectives
3. Materials and Methods
3.1. General Experimental
3.2. Sponge Material
3.3. Extraction and Isolation
3.4. Compound Data
3.5. Structural Similarity Networking
3.6. Molecular Modeling
3.6.1. Acquisition of Target Compound Structures
3.6.2. Acquisition of Target Protein Structures
3.6.3. Molecular Docking between Compounds and Target Proteins
3.7. Data Dependent UHPLC-HRMS/MS Analysis
3.8. Molecular Network Analysis
3.9. Zebrafish Assays
3.9.1. Zebrafish Larvae Assay Procedures
3.9.2. Zebrafish Breeding and Growth
3.9.3. Behavioral Data Analysis
3.9.4. Statistical Analysis
3.10. Antimicrobial Activity Testing
3.11. Anthelmintic Activity Testing
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 4 April 2022).
- Miguel, C.P.V.; Mejias, A.; Leber, A.; Sanchez, P.J. A decade of antimicrobial resistance in Staphylococcus aureus: A single center experience. PLoS ONE 2019, 14, e0212029. [Google Scholar]
- Craft, K.M.; Nguyen, J.M.; Berg, L.J.; Townsend, S.D. Methicillin-resistant Staphylococcus aureus (MRSA): Antibiotic-Resistance and the Biofilm Phenotype. MedChemComm 2019, 10, 1231–1241. [Google Scholar] [CrossRef]
- Mayer, A.; Guerrero, A.J.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Nakamura, F.; Fusetani, N. Marine pharmacology in 2014–2015: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, antiviral, and anthelmintic activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2020, 18, 5. [Google Scholar]
- Mayer, A.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2009–2011: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2013, 11, 2510–2573. [Google Scholar] [PubMed]
- Mayer, A.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Fusetani, N. Marine pharmacology in 2012–2013: Marine compounds with antibacterial, antidiabetic, antifungal, anti-inflammatory, antiprotozoal, antituberculosis, and antiviral activities; affecting the immune and nervous systems, and other miscellaneous mechanisms of action. Mar. Drugs 2017, 15, 273. [Google Scholar]
- Mayer, A.M.S.; Guerrero, A.J.; Rodríguez, A.D.; Taglialatela-Scafati, O.; Nakamura, F.; Fusetani, N. Marine Pharmacology in 2016–2017: Marine Compounds with Antibacterial, Antidiabetic, Antifungal, Anti-Inflammatory, Antiprotozoal, Antituberculosis and Antiviral Activities; Affecting the Immune and Nervous Systems, and Other Miscellaneous Mechanisms of Action. Mar. Drugs 2021, 19, 49. [Google Scholar] [PubMed]
- Liu, M.; El-Hossary, E.M.; A Oelschlaeger, T.; Donia, M.S.; Quinn, R.J.; Abdelmohsen, U.R. Potential of marine natural products against drug-resistant bacterial infections. Lancet Infect. Dis. 2019, 19, e237–e245. [Google Scholar] [CrossRef]
- Lever, J.; Brkljača, R.; Rix, C.; Urban, S. Application of Networking Approaches to Assess the Chemical Diversity, Biogeography, and Pharmaceutical Potential of Verongiida Natural Products. Mar. Drugs 2021, 19, 582. [Google Scholar] [CrossRef]
- Xu, M.; Davis, R.A.; Feng, Y.; Sykes, M.L.; Shelper, T.; Avery, V.M.; Camp, D.; Quinn, R.J. Ianthelliformisamines A–C, antibacterial bromotyrosine-derived metabolites from the marine sponge Suberea ianthelliformis. J. Nat. Prod. 2012, 75, 1001–1005. [Google Scholar] [CrossRef]
- Pieri, C.; Borselli, D.; Di Giorgio, C.; De Méo, M.; Bolla, J.-M.; Vidal, N.; Combes, S.; Brunel, J.M. New ianthelliformisamine derivatives as antibiotic enhancers against resistant Gram-negative bacteria. J. Med. Chem. 2014, 57, 4263–4272. [Google Scholar] [CrossRef]
- Thoms, C.; Schupp, P.J. Activated chemical defense in marine sponges—A case study on Aplysinella rhax. J. Chem. Ecol. 2008, 34, 1242–1252. [Google Scholar] [CrossRef] [PubMed]
- Thoms, C.; Wolff, M.; Padmakumar, K.; Ebel, R.; Proksch, P. Chemical defense of Mediterranean sponges Aplysina cavernicola and Aplysina aerophoba. Z. Naturforsch. 2004, 59, 113–122. [Google Scholar] [CrossRef]
- Ortlepp, S.; Sjögren, M.; Dahlström, M.; Weber, H.; Ebel, R.; Edrada, R.; Thoms, C.; Schupp, P.; Bohlin, L.; Proksch, P. Antifouling activity of bromotyrosine-derived sponge metabolites and synthetic analogues. Mar. Biotechnol. 2007, 9, 776–785. [Google Scholar] [CrossRef]
- Teeyapant, R.; Woerdenbag, H.J.; Kreis, P.; Hacker, J.; Wray, V.; Witte, L.; Proksch, P. Antibiotic and cytotoxic activity of brominated compounds from the marine sponge Verongia aerophoba. Z. Naturforsch. 1993, 48, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Teeyapant, R.; Proksch, P. Biotransformation of brominated compounds in the marine sponge Verongia aerophoba—Evidence for an induced chemical defense? Sci. Nat. 1993, 80, 369–370. [Google Scholar] [CrossRef]
- Teeyapant, R.; Kreis, P.; Wray, V.; Witte, L.; Proksch, P. Brominated secondary compounds from the marine sponge Verongia aerophoba and the sponge feeding gastropod Tylodina perversa. Z. Naturforsch. 1993, 48, 640–644. [Google Scholar] [CrossRef]
- Kumar, M.S.L.; Ali, K.; Chaturvedi, P.; Meena, S.; Datta, D.; Panda, G. Design, synthesis and biological evaluation of oxime lacking Psammaplin inspired chemical libraries as anti-cancer agents. J. Mol. Struct. 2021, 1225, 129173. [Google Scholar] [CrossRef]
- Jing, Q.; Hu, X.; Ma, Y.; Mu, J.; Liu, W.; Xu, F.; Li, Z.; Bai, J.; Hua, H.; Li, D. Marine-Derived Natural Lead Compound Disulfide-Linked Dimer Psammaplin A: Biological Activity and Structural Modification. Mar. Drugs 2019, 17, 384. [Google Scholar] [CrossRef]
- Bao, Y.; Xu, Q.; Wang, L.; Wei, Y.; Hu, B.; Wang, J.; Liu, D.; Zhao, L.; Jing, Y. Studying Histone Deacetylase Inhibition and Apoptosis Induction of Psammaplin A Monomers with Modified Thiol Group. ACS Med. Chem. Lett. 2021, 12, 39–47. [Google Scholar] [CrossRef]
- Kaur, K.; Kumar, V.; Sharma, A.K.; Gupta, G.K. Isoxazoline containing natural products as anticancer agents: A review. Eur. J. Med. Chem. 2014, 77, 121–133. [Google Scholar] [CrossRef]
- Peng, J.; Li, J.; Hamann, M.T. The Marine Bromotyrosine Derivatives. Alkaloids Chem. Biol. 2005, 61, 59–262. [Google Scholar] [PubMed]
- Bechelane-Maia, E.H.; Assis, L.C.; Alves de Oliveira, T.; Marques da Silva, A.; Gutterres Taranto, A. Structure-based virtual screening: From classical to artificial intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Fox Ramos, A.E.; Evanno, L.; Poupon, E.; Champy, P.; Beniddir, M.A. Natural products targeting strategies involving molecular networking: Different manners, one goal. Nat. Prod. Rep. 2019, 36, 960–980. [Google Scholar] [CrossRef]
- Quinn, R.A.; Nothias, L.-F.; Vining, O.; Meehan, M.; Esquenazi, E.; Dorrestein, P.C. Molecular Networking As a Drug Discovery, Drug Metabolism, and Precision Medicine Strategy. Trends Pharmacol. Sci. 2017, 38, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Abou-Shoer, M.I.; Shaala, L.A.; Youssef, D.T.A.; Badr, J.M.; Habib, A.-A.M. Bioactive brominated metabolites from the Red Sea sponge Suberea mollis. J. Nat. Prod. 2008, 71, 1464–1467. [Google Scholar] [CrossRef]
- Kernan, M.R.; Cambie, R.C. Chemistry of sponges, VII: 11, 19-dideoxyfistularin 3 and 11-hydroxyaerothionin, bromotyrosine derivatives from Pseudoceratina durissima. J. Nat. Prod. 1990, 53, 615–622. [Google Scholar] [CrossRef]
- Qi, S.-H.; Wang, Y.-F.; Zhang, S. Steroids and alkaloids from the South China Sea sponge Axinella sp. J. Asian Nat. Prod. Res. 2009, 11, 1040–1044. [Google Scholar] [CrossRef]
- Huang, X.-P.; Deng, Z.-W.; van Soest, R.W.M.; Lin, W.-H. Brominated derivatives from the Chinese sponge Pseudoceratina sp. J. Asian Nat. Prod. Res. 2008, 10, 239–242. [Google Scholar] [CrossRef]
- Lipowicz, B.; Hanekop, N.; Schmitt, L.; Proksch, P. An aeroplysinin-1 specific nitrile hydratase isolated from the marine sponge Aplysina cavernicola. Mar. Drugs 2013, 11, 3046–3067. [Google Scholar] [CrossRef]
- Shaala, L.A.; Khalifa, S.I.; Mesbah, M.K.; van Soest, R.W.M.; Youssef, D.T.A. Subereaphenol A, a new cytotoxic and antimicrobial dibrominated phenol from the Red Sea sponge Suberea mollis. Nat. Prod. Comm. 2008, 3, 219–222. [Google Scholar] [CrossRef]
- Longeon, A.; Guyot, M.; Vacelet, J. Araplysillins-I and-II: Biologically active dibromotyrosine derivatives from the sponge Psammaplysilla arabica. Experientia 1990, 46, 548–550. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A.; Badr, J.M.; Sulaiman, M.; Khedr, A. Bioactive secondary metabolites from the Red Sea marine Verongid sponge Suberea species. Mar. Drugs 2015, 13, 1621–1631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- OSIRIS Property Explorer. 2021. Available online: https://www.organic-chemistry.org/prog/peo/ (accessed on 3 March 2021).
- Sayed, A.M.; Alhadrami, H.A.; El-Hawary, S.S.; Mohammed, R.; Hassan, H.M.; Rateb, M.E.; Abdelmohsen, U.R.; Bakeer, W. Discovery of Two Brominated Oxindole Alkaloids as Staphylococcal DNA Gyrase and Pyruvate Kinase Inhibitors via Inverse Virtual Screening. Microorganisms 2020, 8, 293. [Google Scholar] [CrossRef]
- Zoraghi, R.; Worrall, L.; See, R.H.; Strangman, W.; Popplewell, W.L.; Gong, H.; Samaai, T.; Swayze, R.D.; Kaur, S.; Vuckovic, M.; et al. Methicillin-resistant Staphylococcus aureus (MRSA) pyruvate kinase as a target for bis-indole alkaloids with antibacterial activities. J. Biol. Chem. 2011, 286, 44716–44725. [Google Scholar] [CrossRef]
- Chan, B.C.; Ip, M.; Lau, C.; Lui, S.; Jolivalt, C.; Ganem-Elbaz, C.; Litaudon, M.; Reiner, N.E.; Gong, H.; See, R.H.; et al. Synergistic effects of baicalein with ciprofloxacin against NorA over-expressed methicillin-resistant Staphylococcus aureus (MRSA) and inhibition of MRSA pyruvate kinase. J. Ethnopharmacol. 2011, 137, 767–773. [Google Scholar] [CrossRef]
- Lade, H.; Kim, J.S. Bacterial Targets of Antibiotics in Methicillin-Resistant Staphylococcus aureus. Antibiotics 2021, 10, 398. [Google Scholar] [CrossRef]
- Ohlsen, K.; Lorenz, U. Novel targets for antibiotics in Staphylococcus aureus. Fut. Med. 2007, 2, 655–666. [Google Scholar] [CrossRef]
- Khoshnood, S.; Heidary, M.; Asadi, A.; Soleimani, S.; Motahar, M.; Savari, M.; Saki, M.; Abdi, M. A review on mechanism of action, resistance, synergism, and clinical implications of mupirocin against Staphylococcus aureus. Biomed. Pharmacother. 2019, 109, 1809–1818. [Google Scholar] [CrossRef]
- Torrelo, A.; Grimalt, R.; Masramon, X.; López, N.A.; Zsolt, I. Ozenoxacin, a new effective and safe topical treatment for impetigo in children and adolescents. Dermatology 2020, 236, 199–207. [Google Scholar] [CrossRef]
- Benson, T.E.; Harris, M.S.; Choi, G.H.; Cialdella, J.I.; Herberg, J.T.; Martin, J.P.; Baldwin, E.T. A structural variation for MurB: X-ray crystal structure of Staphylococcus aureus UDP-N-acetylenolpyruvylglucosamine reductase (MurB). Biochemistry 2001, 40, 2340–2350. [Google Scholar] [CrossRef] [PubMed]
- Nakama, T.; Nureki, O.; Yokoyama, S. Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J. Biol. Chem. 2001, 276, 47387–47393. [Google Scholar] [CrossRef]
- Torres-Larios, A.; Sankaranarayanan, R.; Rees, B.; Dock-Bregeon, A.-C.; Moras, D. Conformational Movements and Cooperativity upon Amino Acid, ATP and tRNA Binding in Threonyl-tRNA Synthetase. J. Mol. Biol. 2003, 331, 201–211. [Google Scholar] [CrossRef]
- Yoon, H.J.; Kim, H.L.; Lee, S.-K.; Kim, H.-W.; Lee, J.Y.; Mikami, B.; Suh, S.W. Crystal structure of peptide deformylase from Staphylococcus aureus in complex with actinonin, a naturally occurring antibacterial agent. Proteins 2004, 57, 639–642. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chang, J.S.; Herberg, J.T.; Horng, M.-M.; Tomich, P.K.; Lin, A.H.; Marotti, K.R. Allosteric inhibition of Staphylococcus aureus d-alanine: D-alanine ligase revealed by crystallographic studies. Proc. Natl. Acad. Sci. USA 2006, 103, 15178–15183. [Google Scholar] [CrossRef]
- Heaslet, H.; Harris, M.; Fahnoe, K.; Sarver, R.; Putz, H.; Chang, J.; Subramanyam, C.; Barreiro, G.; Miller, J.R. Structural comparison of chromosomal and exogenous dihydrofolate reductase from Staphylococcus aureus in complex with the potent inhibitor trimethoprim. Proteins Struct. Funct. Bioinf. 2009, 76, 706–717. [Google Scholar] [CrossRef]
- Bax, B.D.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Type IIA topoisomerase inhibition by a new class of antibacterial agents. Nature 2010, 466, 935–940. [Google Scholar] [CrossRef]
- Ronkin, S.M.; Badia, M.; Bellon, S.; Grillot, A.-L.; Gross, C.H.; Grossman, T.H.; Mani, N.; Parsons, J.D.; Stamos, D.; Trudeau, M.; et al. Discovery of pyrazolthiazoles as novel and potent inhibitors of bacterial gyrase. Bioorg. Med. Chem. Lett. 2010, 20, 2828–2831. [Google Scholar] [CrossRef]
- Axerio-Cilies, P.; Chan, P.F.; Eggleston, D.S.; Fosberry, A.; Gentry, D.R.; Gorrec, F.; Giordano, I.; Hann, M.M.; Hennessy, A.; Hibbs, M.; et al. Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase. ACS Chem. Biol. 2012, 7, 350–359. [Google Scholar] [CrossRef]
- Matsui, T.; Yamane, J.; Mogi, N.; Yamaguchi, H.; Takemoto, H.; Yao, M.; Tanaka, I. Structural reorganization of the bacterial cell-division protein FtsZ from Staphylococcus aureus. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68 Pt 9, 1175–1188. [Google Scholar] [CrossRef]
- Schiebel, J.; Chang, A.; Shah, S.; Lu, Y.; Liu, L.; Pan, P.; Hirschbeck, M.W.; Tareilus, M.; Eltschkner, S.; Yu, W.; et al. Rational design of broad spectrum antibacterial activity based on a clinically relevant enoyl-acyl carrier protein (ACP) reductase inhibitor. J. Biol. Chem. 2014, 289, 15987–16005. [Google Scholar] [CrossRef] [PubMed]
- Boundy, S.; Safo, M.K.; Wang, L.; Musayev, F.N.; O’Farrell, H.C.; Rife, J.P.; Archer, G.L. Characterization of the Staphylococcus aureus rRNA methyltransferase encoded by orfX, the gene containing the staphylococcal chromosome Cassette mec (SCCmec) insertion site. J. Biol. Chem. 2013, 288, 132–140. [Google Scholar] [CrossRef]
- Surivet, J.P.; Lange, R.; Hubschwerlen, C.; Keck, W.; Specklin, J.-L.; Ritz, D.; Bur, D.; Locher, H.; Seiler, P.; Strasser, D.S.; et al. Structure-guided design, synthesis and biological evaluation of novel DNA ligase inhibitors with in vitro and in vivo anti-staphylococcal activity. Bioorg. Med. Chem. Lett. 2012, 22, 6705–6711. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Patel, S.; Sharma, N.; Soisson, S.M.; Kishii, R.; Takei, M.; Fukuda, Y.; Lumb, K.J.; Singh, S.B. Structures of kibdelomycin bound to Staphylococcus aureus GyrB and ParE showed a novel U-shaped binding mode. ACS Chem. Biol. 2014, 9, 2023–2031. [Google Scholar] [CrossRef] [PubMed]
- Ting, Y.T.; Harris, P.W.R.; Batot, G.; Brimble, M.A.; Baker, E.N.; Young, P.G. Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization. IUCrJ 2016, 3 Pt 1, 10–19. [Google Scholar] [CrossRef]
- Kim, T.; Choi, J.; Lee, S.; Yeo, K.J.; Cheong, H.-K.; Kim, K.K. Structural Studies on the Extracellular Domain of Sensor Histidine Kinase YycG from Staphylococcus aureus and Its Functional Implications. J. Mol. Biol. 2016, 428, 3074–3089. [Google Scholar] [CrossRef]
- Mahasenan, K.V.; Molina, R.; Bouley, R.; Batuecas, M.T.; Fisher, J.F.; Hermoso, J.A.; Chang, M.; Mobashery, S. Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis. J. Am. Chem. Soc. 2017, 139, 2102–2110. [Google Scholar] [CrossRef]
- Bemis, G.W.; Murcko, M.A. The properties of known drugs. 1. Molecular frameworks. J. Med. Chem. 1996, 39, 2887–2893. [Google Scholar]
- Blondel, V.D.; Guillaume, J.-L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef]
- Nothias, L.F.; Petras, D.; Schmid, R.; Dührkop, K.; Rainer, J.; Sarvepalli, A.; Protsyuk, I.; Ernst, M.; Tsugawa, H.; Fleischauer, M.; et al. Feature-based molecular networking in the GNPS analysis environment. Nat. Methods 2020, 17, 905–908. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Allard, P.M.; Péresse, T.; Bisson, J.; Gindro, K.; Marcourt, L.; Van Cuong, P.; Roussi, F.; Litaudon, M.; Wolfender, J.-L. Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication. Anal. Chem. 2016, 88, 3317–3323. [Google Scholar] [CrossRef]
- Rutz, A.; Dounoue-Kubo, M.; Ollivier, S.; Bisson, J.; Bagheri, M.; Saesong, T.; Ebrahimi, S.N.; Ingkaninan, K.; Wolfender, J.-L.; Allard, P.-M. Taxonomically informed scoring enhances confidence in natural products annotation. Front. Plant Sci. 2019, 10, 1329. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Matsunaga, S.; Fusetani, N. Purpuramines A-I, new bromotyrosine-derived metabolites from the marine sponge Psammaplysilla purpurea. Tetrahedron 1993, 49, 3749–3754. [Google Scholar] [CrossRef]
- Cárdenas, P. Who Produces Ianthelline? The Arctic Sponge Stryphnus fortis or its Sponge Epibiont Hexadella dedritifera: A Probable Case of Sponge-Sponge Contamination. J. Chem. Ecol. 2016, 42, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Salim, A.A.; Khalil, Z.G.; Capon, R.J. Structural and stereochemical investigations into bromotyrosine-derived metabolites from southern Australian marine sponges, Pseudoceratina spp. Tetrahedron 2012, 68, 9802–9807. [Google Scholar] [CrossRef]
- Kobayashi, J.; Honma, K.; Tsuda, M. Purealidins J-R new bromotyrosine alkaloids from the Okinawan marine sponge Pammplysilla purea. Chem. Pharm. Bull. 1995, 43, 403–407. [Google Scholar] [CrossRef]
- Nicacio, K.J.; Ioca, L.P.; Froes, A.M.; Leomil, L.; Appolinario, L.R.; Thompson, C.C.; Thompson, F.L.; Ferreira, A.G.; Williams, D.E.; Andersen, R.J.; et al. Cultures of the Marine Bacterium Pseudovibrio denitrificans Ab134 Produce Bromotyrosine-Derived Alkaloids Previously Only Isolated from Marine Sponges. J. Nat. Prod. 2017, 80, 235–240. [Google Scholar] [CrossRef]
- Rodriguez, A.D.; Pina, I.C. The structures of aplysinamisines I, II, and III New bromotyrosine-derived alkaloids from the Caribbean sponge Aplysina cauliformi. . J. Nat. Prod. 1993, 56, 907–914. [Google Scholar] [CrossRef]
- Fendert, T.; Wray, V.; van Soest, R.W.M.; Proksch, P. Bromoisoxazoline Alkaloids from the Caribbean Sponge Aplysina insularis. Z. Für Nat. C 1999, 54, 246–252. [Google Scholar] [CrossRef]
- Tilvi, S.; Rodrigues, C.; Naik, C.; Parameswaran, P.; Wahidhulla, S. New bromotyrosine alkaloids from the marine sponge Psammaplysilla purpurea. Tetrahedron 2004, 60, 10207–10215. [Google Scholar] [CrossRef]
- Tilvi, S.; Majik, M. 2D NMR Studies of Bromotyrosine Alkaloid, Purpurealidin K from Marine Sponge Psammaplysilla purpurea. ChemistrySelect 2019, 4, 6568–6571. [Google Scholar] [CrossRef]
- Tran, T.D.; Pham, N.B.; Fechner, G.; Hooper, J.N.A.; Quinn, R.J. Bromotyrosine Alkaloids from the Australian Marine Sponge Pseudoceratina verrucosa. J. Nat. Prod. 2013, 76, 516–523. [Google Scholar] [CrossRef]
- Teruya, T.; Iwasaki, A.; Suenaga, K. 20-N-methylpurpuramine E: New bromotyrosine-derived metabolite from Okinawan marine sponge Pseudoceratina purpurea. Bull. Chem. Soc. Jpn. 2008, 81, 1026–1027. [Google Scholar] [CrossRef]
- Tilvi, S.; D’Souza, L. Identifying the related compounds using electrospray ionization tandem mass spectrometry: Bromotyrosine alkaloids from marine sponge Psammaplysilla purpurea. Eur. J. Mass Spectrom. 2012, 18, 333–343. [Google Scholar] [CrossRef]
- Tian, L.W.; Feng, Y.; Shimizu, Y.; Pfeifer, T.; Wellington, C.; Hooper, J.N.; Quinn, R.J. Aplysinellamides A-C, bromotyrosine-derived metabolites from an Australian Aplysinella sp. marine sponge. J. Nat. Prod. 2014, 77, 1210–1214. [Google Scholar] [CrossRef]
- Weiss, B.; Ebel, R.; Elbrächter, M.; Kirchner, M.; Proksch, P. Defense metabolites from the marine sponge. Verongia Aerophoba. Biochem. Syst. Ecol. 1996, 24, 1–12. [Google Scholar] [CrossRef]
- Badr, J.M.; Shaala, L.A.; Abou-Shoer, M.I.; Tawfik, M.K.; Habib, A.-A.M. Bioactive Brominated Metabolites from the Red Sea Sponge Pseudoceratina arabica. J. Nat. Prod. 2008, 71, 1472–1474. [Google Scholar] [CrossRef]
- Mierzwa, R.; King, A.; Conover, M.A.; Tozzi, S.; Puar, M.S.; Patel, M.; Coval, S.J.; Pomponi, S.A. Verongamine, a Novel Bromotyrosine-Derived Histamine H3-Antagonist from the Marine Sponge Verongula gigantea. J. Nat. Prod. 1994, 57, 175–177. [Google Scholar] [CrossRef]
- Thirionet, I.; Daloze, D.; Braekman, J.C.; Willemsen, P. 5-Bromoverongamine, a Novel Antifouling Tyrosine Alkaloid from the Sponge Pseudoceratina sp. Nat. Prod. Lett. 1998, 12, 209–214. [Google Scholar] [CrossRef]
- Kassuhlke, K.E.; Faulkner, J.D. Two new dibromotyrosine derivatives from the caribbean sponge Pseudoceratina Crassa. Tetrahedron 1991, 47, 1809–1814. [Google Scholar] [CrossRef]
- Manzo, E.; van Soest, R.; Matainaho, L.; Roberge, M.; Anderson, R.J. Ceratamines A and B, antimitotic heterocyclic alkaloids isolated from the marine sponge Pseudoceratina sp collected in Papua New Guinea. Org. Lett. 2003, 5, 4591–4594. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, J.; Honma, K.; Tsuda, M. Lipopurealins D and E and purealidin H new bromotyrosine alkaloids from the okinawan marine sponge Psammplysilla purea. J. Nat. Prod. 1995, 58, 467–470. [Google Scholar] [CrossRef]
- Tsuda, M.; Shigemori, H.; Ishibashi, M.; Kobayashi, J. Purealidins E-G, New Bromotyrosine Alkaloids from the Okinawan Marine Sponge Psammaplysilla purea. J. Nat. Prod. 1992, 55, 1325–1327. [Google Scholar] [CrossRef]
- Mohanty, I.; Tapadar, S.; Moore, S.G.; Biggs, J.S.; Freeman, C.J.; Gaul, D.A.; Garg, N.; Agarwal, V. Presence of bromotyrosine alkaloids in marine sponges is independent of metabolic and microbiome architectures. Msystems 2021, 6, 1–17. [Google Scholar] [CrossRef]
- Tsukamoto, S.; Kato, H.; Hirota, H.; Fusetani, N. Ceratinamine: An Unprecedented Antifouling Cyanoformamide from the Marine Sponge Pseudoceratina purpurea. J. Org. Chem. 1996, 61, 2936–2937. [Google Scholar] [CrossRef]
- Acosta, A.I.; Rodriguez, A.D. 11-Oxoaerothionin A Cytotoxic Antitumor Bromotyrosine-Derived Alkaloid from the Caribbean Marine Sponge Aplysina lacunosa. J. Nat. Prod. 1992, 55, 1007–1012. [Google Scholar] [CrossRef]
- Rogers, E.W.; Molinski, T.F. Highly Polar Spiroisoxazolines from the Sponge Aplysina fulva. J. Nat. Prod. 2007, 70, 1191–1194. [Google Scholar] [CrossRef]
- Xynas, R.; Capon, R.J. Two New Bromotyrosine-Derived Metabolites From an Australian Marine Sponge, Aplysina sp. Aust. J. Chem. 1989, 42, 1427–1433. [Google Scholar] [CrossRef]
- Rama Roa, M.; Venkatesham, U.; Venkateswarlu, Y. Two bromo compounds from the sponge Psammaplysilla purpurea. Ind. J. Chem. 1999, 38B, 1301–1303. [Google Scholar]
- Goud, T.V.; Srinivasulu, M.; Reddy, V.L.N.; Reddy, A.V.; Rao, T.P.; Kumar, D.S.; Murty, U.S.; Venkateswarlu, Y. Two New Bromotyrosine-Derived Metabolites from the Sponge Psammaplysilla purpurea. Chem. Pharm. Bull. 2003, 51, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.W.; Fernanda de Oliveira, M.; Berlinck, R.G.S.; Konig, G.M.; Molinski, T.F. Stereochemical Heterogeneity in Verongid Sponge Metabolites. Absolute Stereochemistry of (+)-Fistularin-3 and (+)-11-epi-Fistularin-3 by Microscale LCMS-Marfey’s Analysis. J. Nat. Prod. 2005, 68, 891–896. [Google Scholar] [CrossRef] [PubMed]
- Kijjoa, A.; Bessa, J.; Wattanadilok, R.; Sawangwong, P.; Nascimento, N.S.J.; Pedro, M.; Silva, A.M.S.; Eaton, G.; van Soest, R.; Herz, W. Dibromotyrosine derivatives, a maleimide, aplysamine-2 and other constituents of the marine sponge Pseudoceratina purpurea. Z. Naturforsch. 2005, 60b, 904–908. [Google Scholar] [CrossRef]
- Compagnone, R.S.; Avila, R.; Suarez, A.I.; Abrams, O.V.; Rangel, H.R.; Arvelo, F.; Pina, I.C.; Merentes, E. 11-Deoxyfistularin-3, a New Cytotoxic Metabolite from the Caribbean Sponge Aplysina fistularis insularis. J. Nat. Prod. 1999, 62, 1443–1444. [Google Scholar] [CrossRef] [PubMed]
- Ragini, K.; Fromont, J.; Piggott, A.M.; Karuso, P. Enantiodivergence in the Biosynthesis of Bromotyrosine Alkaloids from Sponges? J. Nat. Prod. 2017, 80, 215–219. [Google Scholar] [CrossRef]
- Hirano, K.; Kubota, T.; Tsuda, M.; Watanabe, K.; Fromont, J.; Kobayashi, J. Ma’edamines A and B, Cytotoxic bromotyrosine alkaloids with a Uinque 2(1H)Pyrazinone ring from sponge Suberea sp. Tetrahedron 2000, 56, 8107–8110. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A.; Sulaiman, M.; Behery, F.A.; Foudah, A.I.; Sayed, K.A.E. Subereamolline A as a Potent Breast Cancer Migration, Invasion and Proliferation Inhibitor and Bioactive Dibrominated Alkaloids from the Red Sea Sponge Pseudoceratina arabica. Mar. Drugs 2012, 10, 2492–2508. [Google Scholar] [CrossRef]
- MarinLit Database. Available online: https://pubs.rsc.org/marinlit/ (accessed on 2 March 2019).
- James, D.M.; Kunze, H.B.; Faulkner, D.J. Two new brominated tyrosine derivatives from the sponge Druinella purpurea. J. Nat. Prod. 1991, 54, 1137–1140. [Google Scholar] [CrossRef]
- Moody, K.; Thomson, R.H.; Fattorusso, E.; Minale, L.T.; Sodano, G. Aerothionin and homoaerothionin: Two tetrabromo spirocyclohexadienylisoxazoles from Verongia sponges. J. Chem. Soc. Perkin Trans. 1972, 1, 18–24. [Google Scholar] [CrossRef]
- Okamoto, Y.; Ojika, M.; Kato, S.; Sakagami, Y. Ianthesines A–D, Four Novel Dibromotyrosine-Derived Metabolites from a Marine Sponge, Ianthella sp. Tetrahedron 2000, 56, 5813–5818. [Google Scholar] [CrossRef]
- Aiello, A.; Fattorusso, E.; Menna, M.; Pansini, M. Chemistry of Verongida sponges V. Brominated metabolites from the Caribbean sponge Pseudoceratina sp. Biochem. Syst. Ecol. 1995, 23, 377–381. [Google Scholar]
- Fujiwara, T.; Hwang, J.-H.; Kanamoto, A.; Nagai, H.; Takagi, M.; Shin-Ya, K. JBIR-44, a new bromotyrosine compound from a marine sponge Psammaplysilla purpurea. J. Antibiot. 2009, 62, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Nuñez, C.V.; de Almeida, E.V.; Granato, A.C.; Marques, S.O.; Santos, K.O.; Pereira, F.R.; Macedo, M.L.; Ferreira, A.G.; Hajdu, E.; Pinheiro, U.; et al. Chemical variability within the marine sponge Aplysina fulva. Biochem. Syst. Ecol. 2008, 36, 283–296. [Google Scholar] [CrossRef]
- Shaala, L.A.; Bamane, F.H.; Badr, J.M.; Youssef, D.T.A. Brominated arginine-derived alkaloids from the red sea sponge Suberea mollis. J. Nat. Prod. 2011, 74, 1517–1520. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, M.F.; de Oliveira, J.H.; Galetti, F.C.; de Souza, A.O.; Silva, C.L.; Hajdu, E.; Peixinho, S.; Berlinck, R.G. Antimycobacterial brominated metabolites from two species of marine sponges. Planta Med. 2006, 72, 437–441. [Google Scholar] [CrossRef]
- Shaker, K.H.; Zinecker, H.; Ghani, M.A.; Imhoff, J.F.; Schneider, B. Bioactive metabolites from the sponge Suberea sp. Helv. Chim. Acta 2010, 7, 2880–2887. [Google Scholar] [CrossRef]
- Chang, C.W.; Weinheimer, A.J. 2-Hydroxy, 3, 5-dibromo, 4-methoxyphenylacetamide—A dibromotyrosine metabolite from Psammaplysilla purpurea. Tetrahedron Lett. 1977, 18, 4005–4007. [Google Scholar] [CrossRef]
- Fattorusso, E.; Minale, L.; Sodano, G. Aeroplysinin-1, an antibacterial bromo-compound from the sponge Verongia aerophoba. J. Chem. Soc. 1972, 1, 16–18. [Google Scholar] [CrossRef]
- Gomez-Archila, L.G.; Zapata, W.; Galeano, E.; Martínez, A.; Díaz, F.J.; Rugeles, M.T. Bromotyrosine derivatives from marine sponges inhibit the HIV-1 replication in vitro. Vitae 2014, 21, 114–125. [Google Scholar]
- García-Vilas, J.A.; Martínez-Poveda, B.; Quesada, A.R.; Medina, M.A. Aeroplysinin-1, a sponge-derived multi-targeted bioactive marine drug. Mar. Drugs 2016, 14, 1. [Google Scholar] [CrossRef]
- Kreuter, M.; Bernd, A.; Holzmann, H.; Müller-Klieser, W.; Maidhof, A.; Weißmann, N.; Kljajić, Z.; Batel, R.; Schröder, H.C.; Müller, W. Cytostatic activity of aeroplysinin-1 against lymphoma and epithelioma cells. Z. Naturforsch. C 1989, 44, 680–688. [Google Scholar] [CrossRef]
- Kreuter, M.-H.; Leake, R.E.; Rinaldi, F.; Müller-Klieser, W.; Maidhof, A.; Müller, W.E.; Schröder, H.C. Inhibition of intrinsic protein tyrosine kinase activity of EGF-receptor kinase complex from human breast cancer cells by the marine sponge metabolite (+)-aeroplysinin-1. Comp. Biochem. Physiol. B 1990, 97, 151–158. [Google Scholar] [CrossRef]
- Stuhldreier, F.; Kassel, S.; Schumacher, L.; Wesselborg, S.; Proksch, P.; Fritz, G. Pleiotropic effects of spongean alkaloids on mechanisms of cell death, cell cycle progression and DNA damage response (DDR) of acute myeloid leukemia (AML) cells. Cancer Lett. 2015, 361, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Ogungbemi, A.; Leuthold, D.; Scholz, S.; Küster, E. Hypo-or hyperactivity of zebrafish embryos provoked by neuroactive substances: A review on how experimental parameters impact the predictability of behavior changes. Environ. Sci. Eur. 2019, 31, 88. [Google Scholar] [CrossRef]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Wlodkowic, D. Future prospects of accelerating neuroactive drug discovery with high-throughput behavioral phenotyping. Expert Opin. Drug Discov. 2022, 17, 305–308. [Google Scholar] [CrossRef]
- Henry, J.; Wlodkowic, D. Towards high-throughput chemobehavioural phenomics in neuropsychiatric drug discovery. Mar. Drugs 2019, 17, 340. [Google Scholar] [CrossRef] [Green Version]
- El-Demerdash, A.; Moriou, C.; Toullec, J.; Besson, M.; Soulet, S.; Schmitt, N.; Petek, S.; Lecchini, D.; Debitus, C.; Al-Mourabit, A. Bioactive Bromotyrosine-Derived Alkaloids from the Polynesian Sponge Suberea ianthelliformis. Mar. Drugs 2018, 16, 146. [Google Scholar] [CrossRef]
- Sirimangkalakitti, N.; Olatunji, O.J.; Changwichit, K.; Saesong, T.; Chamni, S.; Chanvorachote, P.; Ingkaninan, K.; Plubrukarn, A.; Suwanborirux, K. Bromotyrosine alkaloids with acetylcholinesterase inhibitory activity from the Thai sponge Acanthodendrilla sp. Nat. Prod. Commun. 2015, 10, 1945–1949. [Google Scholar] [CrossRef]
- Olatunji, O.J.; Ogundajo, A.L.; Oladosu, I.A.; Changwichit, K.; Ingkaninan, K.; Yuenyongsawad, S.; Plubrukarn, A. Non-competitive inhibition of acetylcholinesterase by bromotyrosine alkaloids. Nat. Prod. Commun. 2014, 9, 1559–1561. [Google Scholar] [CrossRef]
- Cesário, H.P.S.d.F.; O Silva, F.C.; A Ferreira, M.K.; de Menezes, J.E.S.A.; Dos Santos, H.S.; Nogueira, C.E.S.; de Silva, K.S.B.; Hajdu, E.; Silveira, E.R.; Pessoa, O.D.L. Anxiolytic-like effect of brominated compounds from the marine sponge Aplysina fulva on adult zebrafish (Danio rerio): Involvement of the GABAergic system. Neurochem. Int. 2021, 146, 105021. [Google Scholar] [CrossRef] [PubMed]
- Gotsbacher, M.P.; Karuso, P. New antimicrobial bromotyrosine analogues from the sponge Pseudoceratina purpurea and its predator Tylodina corticalis. Mar. Drugs 2015, 13, 1389–1409. [Google Scholar] [CrossRef] [PubMed]
- Carnovali, M.; Ciavatta, M.L.; Mollo, E.; Roussis, V.; Banfi, G.; Carbone, M.; Mariotti, M. Aerophobin-1 from the marine sponge Aplysina aerophoba modulates osteogenesis in zebrafish larvae. Mar. Drugs 2022, 20, 135. [Google Scholar] [CrossRef] [PubMed]
- Maahury, M.F.; Allo, V.L. The Computational Calculation and Molecular Docking of Aeroplysinin-1 As Antibacterial. Indones. J. Chem. Res. 2021, 9, 124–128. [Google Scholar] [CrossRef]
- Felix, L.; Mylonakis, E.; Fuchs, B.B. Thioredoxin Reductase Is a Valid Target for Antimicrobial Therapeutic Development Against Gram-Positive Bacteria. Front. Microbiol. 2021, 12, 889. [Google Scholar] [CrossRef]
- Uziel, O.; Borovok, I.; Schreiber, R.; Cohen, G.; Aharonowitz, Y. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 2004, 186, 326–334. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.M.; Lee, R.T. Thioredoxin and thioredoxin target proteins: From molecular mechanisms to functional significance. Antioxid. Redox Signal. 2013, 18, 1165–1207. [Google Scholar] [CrossRef] [Green Version]
- Saccoccia, F.; Angelucci, F.; Boumis, G.; Carotti, D.; Desiato, G.; Miele, A.; Bellelli, A. Thioredoxin reductase and its inhibitors. Curr. Protein Pept. Sci. 2014, 15, 621–646. [Google Scholar] [CrossRef]
- Buey, R.M.; Schmitz, R.A.; Buchanan, B.B.; Balsera, M. Crystal Structure of the Apo-Form of NADPH-Dependent Thioredoxin Reductase from a Methane-Producing Archaeon. Antioxidants 2018, 7, 166. [Google Scholar] [CrossRef]
- Sweeney, N.L.; Lipker, L.; Hanson, A.M.; Bohl, C.J.; Engel, K.E.; Kalous, K.S.; Stemper, M.E.; Sem, D.S.; Schwan, W.R. Docking into Mycobacterium tuberculosis thioredoxin reductase protein yields pyrazolone lead molecules for methicillin-resistant Staphylococcus aureus. Antibiotics 2017, 6, 4. [Google Scholar] [CrossRef]
- Wilkens, S.J.; Janes, A.J.; Su, A.I. HierS: Hierarchical Scaffold Clustering using Topological Chemical Graphs. J. Med. Chem. 2005, 48, 3182–3193. [Google Scholar] [CrossRef]
- Kruger, F.; Stiefl, N.; Landrum, G.A. rdScaffoldNetwork: The Scaffold Network Implementation in RDKit. J. Chem. Inf. Model. 2020, 60, 3331–3335. [Google Scholar] [CrossRef] [PubMed]
- Fruchterman, T.M.J.; Reingold, E.M. Graph drawing by Force-directed Placement. Softw. Pract. Exp. 1991, 21, 1129–1164. [Google Scholar] [CrossRef]
- UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, 480–489. [Google Scholar]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Bertoni, M.; Kiefer, F.; Biasini, M.; Bordoli, L.; Schwede, T. Modeling protein quaternary structure of homo-and hetero-oligomers beyond binary interactions by homology. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef]
- Dallakyan, S.; Olson, A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015, 1263, 243–250. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Henry, J.; Rodriguez, A.; Wlodkowic, D. Impact of digital video analytics on accuracy of chemobehavioural phenotyping in aquatic toxicology. PeerJ 2019, 7, e7367. [Google Scholar] [CrossRef]
- Henry, J.; Bai, Y.; Kreuder, F.; Mawdsley, D.; Kaslin, J.; Wlodkowic, D. Accelerating Chemobehavioral Phenotypic Screening in Neurotoxicology Using a Living Embryo Array System. Zebrafish 2022, 19, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Douek, A.M.; Khabooshan, M.A.; Henry, J.; Stamatis, S.-A.; Kreuder, F.; Ramm, G.; Änkö, M.-L.; Wlodkowic, D.; Kaslin, J. An Engineered sgsh Mutant Zebrafish Recapitulates Molecular and Behavioural Pathobiology of Sanfilippo Syndrome A/MPS IIIA. Int. J. Mol. Sci. 2021, 22, 5948. [Google Scholar] [CrossRef]
- Walpitagama, M.; Carve, M.; Douek, A.; Trestrail, C.; Bai, Y.; Kaslin, J.; Wlodkowic, D. Additives migrating from 3D-printed plastic induce developmental toxicity and neuro-behavioural alterations in early life zebrafish (Danio rerio). Aquat. Toxicol. 2019, 213, 105227. [Google Scholar] [CrossRef] [PubMed]
- Westerfield, M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish. 2000. Available online: http://zfin.org/zf_info/zfbook/zfbk.html (accessed on 10 January 2022).
- Blaskovich, M.A.; Zuegg, J.; Elliott, A.G.; Cooper, M.A. Helping chemists discover new antibiotics. ACS Infect. Dis. 2015, 1, 285–287. [Google Scholar] [CrossRef]
- Taki, A.C.; Byrne, J.; Wang, T.; Sleebs, B.; Nguyen, N.; Hall, R.; Korhonen, P.; Chang, B.; Jackson, P.; Jabbar, A.; et al. High-throughput phenotypic assay to screen for anthelmintic activity on Haemonchus contortus. Pharmaceuticals 2021, 14, 616. [Google Scholar] [CrossRef] [PubMed]
- Preston, S.; Jabbar, A.; Nowell, C.; Joachim, A.; Ruttkowski, B.; Baell, J.; Cardno, T.; Korhonen, P.K.; Piedrafita, D.; Ansell, B.R.; et al. Low cost whole-organism screening of compounds for anthelmintic activity. Int. J. Parasitol. 2015, 45, 333–343. [Google Scholar] [CrossRef] [Green Version]










| Target name | PDBID | MOA | Reference |
|---|---|---|---|
| UDP-N-acetylglucosamine-enolpyruvyl reductase (MurB) | 1hsk | Peptidoglycan synthesis | [43] |
| Isoleucyl-tRNA synthetase | 1jzs | Protein modification | [44] |
| Threonyl-tRNA synthetase | 1nyq | Protein modification | [45] |
| Peptide deformylase (Pdf) | 1q1y | Protein modification | [46] |
| B-ketoacyl-synthase I/II (FabF) | 2gqd | Fatty acid synthesis | To be published |
| D-alanine ligase (Ddl) | 2i87 | Peptidoglycan synthesis | [47] |
| Dihydrofolate reductase (DHFR) | 2w9h | Cellular regulation | [48] |
| DNA Gyrase subunit A (GyrA) | 2xct | DNA replication | [49] |
| DNA Gyrase subunit B (GyrB) | 3g7b | DNA replication | [50] |
| Pyruvate Kinase (PK) | 3t0t | Glycolysis | [51] |
| Filamenting Temperature sensitive mutant Z (Ftsz) | 3vob | Cell division | [52] |
| Enoly-acyl-carrier protein reductase (FabI) | 4cv1 | Fatty acid synthesis | [53] |
| rRNA methyltransferase | 4fak | Protein modification | [54] |
| Thioredoxin reductase (TrxB) | 4gcm | Stress response | To be published |
| DNA Ligase (LigA) | 4glx | Stress response | [55] |
| DNA Topoisomerase IV subunit B (ParE) | 4urn | DNA replication | [56] |
| Signal Peptidase (SpsB) | 4wvj | Protein secretion | [57] |
| Histidine Kinase (YycG/YycF) | 5is1 | Cellular regulation | [58] |
| Peptidoglycan glycosyl transferase (PBP2a) | 5m18 | Peptidoglycan synthesis | [59] |
| Structure | Scaffold Class (Cluster) | Cluster Population (n) | No. of Lipinski/Veber Failures + | OSIRIS Drug Score * |
|---|---|---|---|---|
 |  | 37 | 0 | 0.44 |
 |  | 37 | 1 (MW) | 0.48 |
 |  | 24 | 0 | 0.26 |
 |  | 22 | 0 | 0.21 |
 |  | 22 | 1(MW) | 0.11 |
 |  | 17 | 0 | 0.13 |
 |  | 15 | 0 | 0.17 |
 |  | 13 | 1(MW) | 0.22 |
 |  | 13 | 5(MW, nHBDon, nHBAcc, nRotB, TPSA) | 0.14 |
| Compound | Cytotoxicity 1 (CC50) µg/mL | Haemolysis 2 (HC10) µg/mL |
|---|---|---|
| 1 | >32 | >32 |
| 2 | >32 | >32 |
| 3 | >32 | >32 |
| 5 | >32 | >32 |
| 6 | >32 | >32 |
| 7 | >32 | >32 |
| 8 | >32 | >32 |
| 9 | >32 | 1.032 |
| 10 | >32 | >32 |
| 11 | >32 | >32 |
| 12 | >32 | >32 |
| Positive control | Tamoxifen: 9 | Melittin: 2.7 |
| Compound | MIC (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|
| MRSA 1 | E. coli 2 | K. pneumoniae 3 | A. baumannii 4 | P. aeruginosa 5 | C. albicans 6 | C. neoformans 7 | |
| 1 % inhibition | 32 | 32 | >32 | >32 | >32 | >32 | >32 |
| 95.9, 96.7 | 93.7, 95.2 | −2.8, −3.3 | 21.5, 52.5 | 10.9, 18.2 | 9.5, 9.8 | −6.6, 0.0 | |
| 2 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | −1.5, 6.1 | 0.9, 2.3 | 10.4, 10.9 | −5.2, 11.6 | −2.4, 12.1 | 0.5, 4.2 | −9.1, 11.7 |
| 3 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | −5.9, 7.4 | −2.9, −3.1 | 10.2, 6.6 | 13.8, 9.8 | 10.4, 2.2 | 3.4, 6.3 | 10.6, 16.1 |
| 5 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 10.0, 19.8 | 8.1, 9.5 | 6.5, 8.7 | 10.2, 8.5 | 4.3, 7.2 | 2.5, 3.3 | 10.4, 3.8 |
| 6 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 3.3, 7.0 | 14.6, 15.6 | 12.3, 6.4 | 2.3, 9.7 | 1.1, 9.7 | 0.6, 4.6 | 11.1, 2.0 |
| 7 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 12.9, 2.5 | 11.2, 6.2 | 10.6, 13.3 | 12.0, 27.4 | 0.1, 13.2 | 1.0, 11.2 | 1.8, 14.1 |
| 8 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 7.6, 8.5 | 12.7, 16.9 | 10.3, 12.5 | 11.0, 7.5 | 11.2, 6.8 | 1.6, 14.3 | 11.1, 4.0 |
| 9 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 0.9, 13.3 | 1.5, 9.6 | 15.5, 6.6 | 10.8, 18.5 | −1.0, 9.2 | −1.2, 12.8 | 11.2, 11.9 |
| 10 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 14.9, 4.7 | 15.8, 6.4 | 6.1, 9.3 | −1.4, −9.2 | −4.3, 10.7 | −0.8, 13.6 | 12.4, 8.5 |
| 11 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 11.3, 5.7 | 15.9, 8.5 | 12.2, 17.0 | 3.4, 7.3 | 0.3, 8.0 | 0.6, 16.2 | 7.6, 7.8 |
| 12 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| % inhibition | 0.4, 8.2 | 12.3, 6.4 | 14.0, 5.6 | −2.1, 12.7 | 17.2, 6.3 | −3.2, 9.2 | −3.4, 8.5 |
| Positive Control | Vancomycin: 1 | Colistin: 0.125 | Colistin: 0.25 | Colistin: 0.25 | Colistin: 0.25 | Fluconazole: 0.125 | Fluconazole: 8 |
| Target Name | PDBID | (+)-Aeroplysinin-1 (1) Affinity (kcal/mol) |
|---|---|---|
| Thioredoxin reductase (TrxB) | 4gcm | −7.0 |
| Signal Peptidase (SpsB) | 4wvj | −6.8 |
| Dihydrofolate reductase (DHFR) | 2w9h | −6.7 |
| UDP-N-acetylglucosamine-enolpyruvyl reductase (MurB) | 1hsk | −6.5 |
| Peptide deformylase (Pdf) | 1q1y | −6.3 |
| Enoly-acyl-carrier protein reductase (FabI) | 4cv1 | −6.3 |
| Threonyl-tRNA synthetase | 1nyq | −6.2 |
| DNA Ligase (LigA) | 4glx | −6.2 |
| Isoleucyl-tRNA synthetase | 1jzs | −6.1 |
| Filamenting Temperature sensitive mutant Z (Ftsz) | 3vob | −5.8 |
| rRNA methyltransferase | 4fak | −5.8 |
| DNA Gyrase subunit A (GyrA) | 2xct | −5.8 |
| Peptidoglycan glycosyl transferase (PBP2a) | 5m18 | −5.7 |
| Pyruvate Kinase (PK) | 3t0t | −5.6 |
| DNA Topoisomerase IV subunit B (ParE) | 4urn | −5.6 |
| B-ketoacyl-synthase I/II (FabF) | 2gqd | −5.6 |
| D-alanine ligase (Ddl) | 2i87 | −5.5 |
| DNA Gyrase subunit B (GyrB) | 3g7b | −5.1 |
| Histidine Kinase (YycG/YycF) | 5is1 | −4.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lever, J.; Kreuder, F.; Henry, J.; Hung, A.; Allard, P.-M.; Brkljača, R.; Rix, C.; Taki, A.C.; Gasser, R.B.; Kaslin, J.; et al. Targeted Isolation of Antibiotic Brominated Alkaloids from the Marine Sponge Pseudoceratina durissima Using Virtual Screening and Molecular Networking. Mar. Drugs 2022, 20, 554. https://doi.org/10.3390/md20090554
Lever J, Kreuder F, Henry J, Hung A, Allard P-M, Brkljača R, Rix C, Taki AC, Gasser RB, Kaslin J, et al. Targeted Isolation of Antibiotic Brominated Alkaloids from the Marine Sponge Pseudoceratina durissima Using Virtual Screening and Molecular Networking. Marine Drugs. 2022; 20(9):554. https://doi.org/10.3390/md20090554
Chicago/Turabian StyleLever, James, Florian Kreuder, Jason Henry, Andrew Hung, Pierre-Marie Allard, Robert Brkljača, Colin Rix, Aya C. Taki, Robin B. Gasser, Jan Kaslin, and et al. 2022. "Targeted Isolation of Antibiotic Brominated Alkaloids from the Marine Sponge Pseudoceratina durissima Using Virtual Screening and Molecular Networking" Marine Drugs 20, no. 9: 554. https://doi.org/10.3390/md20090554