Novel Alkaloids from Marine Actinobacteria: Discovery and Characterization
Abstract
:1. Introduction
2. Novel Alkaloids from Marine Actinobacteria
2.1. Piperidines
2.2. Glutarimides
2.3. Indolizidines
2.4. Pyrroles
2.5. Pyrrolidines
2.6. Pyridines
2.7. Imidazoles
2.8. Pyrimidines
2.9. Indoles
2.9.1. Single Indoles
2.9.2. Bisindoles
2.9.3. Indolocarbazoles
2.10. Diketopiperazines
2.11. Quinolines
2.12. Isoquinolines
2.13. Hybrid Alkaloids
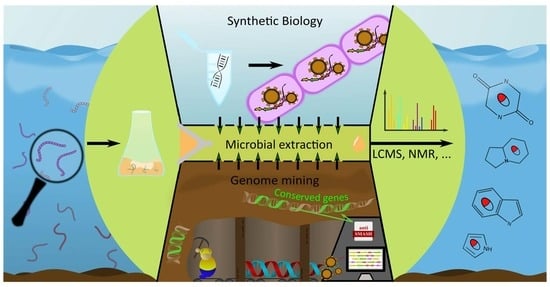
3. Alkaloid Discovery Strategies
3.1. Wet-Lab Strategies
3.1.1. Culturing Strategies
3.1.2. Extraction and Isolation Strategies
3.1.3. Compound Identification
3.2. Genome Mining
3.3. Synthetic Biology in Alkaloid Discovery
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural Products in Drug Discovery: Advances and Opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef] [PubMed]
- Feher, M.; Schmidt, J.M. Property Distributions: Differences between Drugs, Natural Products, and Molecules from Combinatorial Chemistry. J. Chem. Inf. Comput. Sci. 2003, 43, 218–227. [Google Scholar] [CrossRef]
- Montaser, R.; Luesch, H. Marine Natural Products: A New Wave of Drugs? Future Med. Chem. 2011, 3, 1475–1489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigwart, J.D.; Blasiak, R.; Jaspars, M.; Jouffray, J.-B.; Tasdemir, D. Unlocking the Potential of Marine Biodiscovery. Nat. Prod. Rep. 2021, 38, 1235–1242. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine Natural Products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Lu, W.-Y.; Li, H.-J.; Li, Q.-Y.; Wu, Y.-C. Application of Marine Natural Products in Drug Research. Bioorg. Med. Chem. 2021, 35, 116058. [Google Scholar] [CrossRef]
- Munekata, P.E.S.; Pateiro, M.; Conte-Junior, C.A.; Domínguez, R.; Nawaz, A.; Walayat, N.; Fierro, E.M.; Lorenzo, J.M. Marine Alkaloids: Compounds with In Vivo Activity and Chemical Synthesis. Mar. Drugs 2021, 19, 374. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, L.; Zhou, Y.; Han, B. Natural Products from Actinomycetes Associated with Marine Organisms. Mar. Drugs 2021, 19, 629. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Du, W.; Lu, H.; Lan, J.; Liang, K.; Cao, S. A Review: Halogenated Compounds from Marine Actinomycetes. Molecules 2021, 26, 2754. [Google Scholar] [CrossRef]
- Goel, N.; Fatima, W.S.; Kumar, S.; Sinha, R.; Khare, S.K. Antimicrobial Resistance in Biofilms: Exploring Marine Actinobacteria as a Potential Source of Antibiotics and Biofilm Inhibitors. Biotechnol. Rep. 2021, 30, e00613. [Google Scholar] [CrossRef]
- Willems, T.; de Mol, M.L.; de Bruycker, A.; de Maeseneire, S.L.; Soetaert, W.K. Alkaloids from Marine Fungi: Promising Antimicrobials. Antibiotics 2020, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Hopwood, D.A. Natural Product Biosynthesis by Microorganisms and Plants, Part C, 1st ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 517, ISBN 9780124046344. [Google Scholar]
- Xu, W.; Gavia, D.J.; Tang, Y. Biosynthesis of Fungal Indole Alkaloids. Nat. Prod. Rep. 2014, 31, 1474–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reddy, P.; Guthridge, K.; Vassiliadis, S.; Hemsworth, J.; Hettiarachchige, I.; Spangenberg, G.; Rochfort, S. Tremorgenic Mycotoxins: Structure Diversity and Biological Activity. Toxins 2019, 11, 302. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.L.; Wang, Y.S.; Liu, C.L.; Zeng, Y.J.; Cheng, P.; Jiao, R.H.; Bao, S.X.; Huang, H.Q.; Tan, R.X.; Ge, H.M. Strepchazolins A and B: Two New Alkaloids from a Marine Streptomyces chartreusis NA02069. Mar. Drugs 2017, 15, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, H.E.; Batista, J.M.; Melo, W.G.P.; de Paula, G.T.; Pupo, M.T. Structure and Absolute Configuration of Secondary Metabolites from Two Strains of Streptomyces chartreusis Associated with Attine Ants. J. Braz. Chem. Soc. 2019, 30, 2672–2680. [Google Scholar] [CrossRef]
- Liu, C.; Yang, C.; Zeng, Y.; Shi, J.; Li, L.; Li, W.; Jiao, R.; Tan, R.; Ge, H. Chartrenoline, a Novel Alkaloid Isolated from a Marine Streptomyces chartreusis NA02069. Chin. Chem. Lett. 2019, 30, 44–46. [Google Scholar] [CrossRef]
- Awodi, U.R.; Ronan, J.L.; Masschelein, J.; de los Santos, E.L.C.; Challis, G.L. Thioester Reduction and Aldehyde Transamination Are Universal Steps in Actinobacterial Polyketide Alkaloid Biosynthesis. Chem. Sci. 2017, 8, 411–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michael, J.P. Indolizidine and Quinolizidine Alkaloids. Nat. Prod. Rep. 2008, 25, 139–165. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, W.; Ge, H.; Zhang, Z.; Wu, B. Bioactive Streptoglutarimides A–J from the Marine-Derived Streptomyces sp. ZZ741. J. Nat. Prod. 2019, 82, 2800–2808. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Miyano, R.; Iwatsuki, M.; Shirahata, T.; Asami, Y.; Kobayashi, Y.; Shiomi, K.; Petersson, G.A.; Takahashi, Y.; Ōmura, S.; et al. The New Iminium Metabolite Produced by Streptomyces griseus OS-3601. J. Antibiot. 2016, 69, 611–615. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, L.; Chai, W.; Lian, X.Y.; Zhang, Z. A Unique Indolizinium Alkaloid Streptopertusacin A and Bioactive Bafilomycins from Marine-Derived Streptomyces sp. HZP-2216E. Phytochemistry 2017, 144, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.-J.; Li, J.-Q.; Zhang, H.-J.; Ding, W.-J.; Ma, Z.-J. Cyclizidine-Type Alkaloids from Streptomyces sp. HNA39. J. Nat. Prod. 2018, 81, 394–399. [Google Scholar] [CrossRef]
- Weber, A.J.; Herskowitz, J.H. Perspectives on ROCK2 as a Therapeutic Target for Alzheimer’s Disease. Front. Cell. Neurosci. 2021, 15, 636017. [Google Scholar] [CrossRef]
- Cheng, X.W.; Li, J.Q.; Jiang, Y.J.; Liu, H.Z.; Huo, C. A New Indolizinium Alkaloid from Marine-Derived Streptomyces sp. HNA39. J. Asian Nat. Prod. Res. 2021, 23, 913–918. [Google Scholar] [CrossRef]
- Seipp, K.; Geske, L.; Opatz, T. Marine Pyrrole Alkaloids. Mar. Drugs 2021, 19, 514. [Google Scholar] [CrossRef]
- Han, X.; Liu, Z.; Zhang, Z.; Zhang, X.; Zhu, T.; Gu, Q.; Li, W.; Che, Q.; Li, D. Geranylpyrrol A and Piericidin F from Streptomyces sp. CHQ-64 ΔrdmF. J. Nat. Prod. 2017, 80, 1684–1687. [Google Scholar] [CrossRef] [PubMed]
- Macherla, V.R.; Liu, J.; Bellows, C.; Teisan, S.; Nicholson, B.; Lam, K.S.; Potts, B.C.M. Glaciapyrroles A, B, and C, Pyrrolosesquiterpenes from a Streptomyces sp. Isolated from an Alaskan Marine Sediment. J. Nat. Prod. 2005, 68, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Braun, D.R.; Chanana, S.; Rajski, S.R.; Bugni, T.S. Phallusialides A−E, Pyrrole-Derived Alkaloids Discovered from a Marine-Derived Micromonospora sp. Bacterium Using MS-Based Metabolomics Approaches. J. Nat. Prod. 2019, 82, 3432. [Google Scholar] [CrossRef]
- Zhang, Y.-M.; Liu, B.-L.; Zheng, X.-H.; Huang, X.-J.; Li, H.-Y.; Zhang, Y.; Zhang, T.-T.; Sun, D.-Y.; Lin, B.-R.; Zhou, G.-X. Anandins A and B, Two Rare Steroidal Alkaloids from a Marine Streptomyces anandii H41-59. Mar. Drugs 2017, 15, 355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, Z.; Huang, X.; Liu, Y.; Liu, Y.; Wang, P.; Liao, S.; Yang, B.; Zhou, X.; Huang, D.; Wang, J. Cytotoxic Anthracycline and Antibacterial Tirandamycin Analogues from a Marine-Derived Streptomyces sp. SCSIO 41399. J. Antibiot. 2018, 72, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, E.A.; Ismail, N.S.M.; Hagras, M.; Refaat, H. Medicinal Attributes of Pyridine Scaffold as Anticancer Targeting Agents. Future J. Pharm. Sci. 2021, 7, 24. [Google Scholar] [CrossRef]
- Chen, M.-H.; Lian, Y.-Y.; Fang, D.-S.; Chen, L.; Jia, J.; Zhang, W.-L.; Lin, R.; Xie, Y.; Bi, H.-K.; Jiang, H. Identification and Antimicrobial Properties of a New Alkaloid Produced by Marine-Derived Verrucosispora sp. FIM06-0036. Nat. Prod. Res. 2019, 35, 4211–4217. [Google Scholar] [CrossRef]
- Rokon, M.; Karim, U.; Harunari, E.; Sharma, A.R.; Oku, N.; Akasaka, K.; Urabe, D.; Sibero, M.T.; Igarashi, Y. Nocarimidazoles C and D, Antimicrobial Alkanoylimidazoles from a Coral-Derived Actinomycete Kocuria sp.: Application of 1 J C,H Coupling Constants for the Unequivocal Determination of Substituted Imidazoles and Stereochemical Diversity of Anteisoalkyl Chains in Microbial Metabolites. Beilstein J. Org. Chem. 2020, 16, 2719–2727. [Google Scholar] [CrossRef]
- Fang, Z.; Chen, S.; Zhu, Y.; Li, J.; Khan, I.; Zhang, Q.; Zhang, C. A New Uridine Derivative and a New Indole Derivative from the Coral-Associated Actinomycete Pseudonocardia sp. SCSIO 11457. Nat. Prod. Res. 2019, 35, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Shelton, J.; Lu, X.; Hollenbaugh, J.A.; Cho, J.H.; Amblard, F.; Schinazi, R.F. Metabolism, Biochemical Actions, and Chemical Synthesis of Anticancer Nucleosides, Nucleotides, and Base Analogs. Chem. Rev. 2016, 116, 14379–14455. [Google Scholar] [CrossRef]
- Shaala, L.A.; Youssef, D.T.A.; Badr, J.M.; Harakeh, S.M.; Genta-Jouve, G. Bioactive Diketopiperazines and Nucleoside Derivatives from a Sponge-Derived Streptomyces Species. Mar. Drugs 2019, 17, 584. [Google Scholar] [CrossRef] [Green Version]
- Netz, N.; Opatz, T. Marine Indole Alkaloids. Mar. Drugs 2015, 13, 4814–4914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, S.; Zhou, T.-T.; Xie, C.-L.; Zhang, G.-Y.; Yang, X.-W. Microindolinone A, a Novel 4,5,6,7-Tetrahydroindole, from the Deep-Sea-Derived Actinomycete Microbacterium sp. MCCC 1A11207. Mar. Drugs 2017, 15, 230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, Q.; Qiao, L.; Han, X.; Liu, Y.; Wang, W.; Gu, Q.; Zhu, T.; Li, D. Anthranosides A−C, Anthranilate Derivatives from a Sponge-Derived Streptomyces sp. CMN-62. Org. Lett. 2018, 20, 5466–5469. [Google Scholar] [CrossRef] [PubMed]
- El-Hawary, S.S.; Sayed, A.M.; Mohammed, R.; Khanfar, M.A.; Rateb, M.E.; Mohammed, T.A.; Hajjar, D.; Hassan, H.M.; Gulder, T.A.M.; Abdelmohsen, U.R. New Pim-1 Kinase Inhibitor From the Co-Culture of Two Sponge-Associated Actinomycetes. Front. Chem. 2018, 6, 538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, W.; Li, Q.; Song, T.; Chen, L.; Li, X.C.; Zhang, Z.; Lian, X.Y. Isolation, Structure Elucidation, and Antibacterial Evaluation of the Metabolites Produced by the Marine-Sourced Streptomyces sp. ZZ820. Tetrahedron 2019, 75, 1186–1193. [Google Scholar] [CrossRef]
- Song, Y.; Yang, J.; Yu, J.; Li, J.; Yuan, J.; Wong, N.K.; Ju, J. Chlorinated Bis-Indole Alkaloids from Deep-Sea Derived Streptomyces sp. SCSIO 11791 with Antibacterial and Cytotoxic Activities. J. Antibiot. 2020, 73, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Salas, J.A.; Méndez, C. Indolocarbazole Antitumour Compounds by Combinatorial Biosynthesis. Curr. Opin. Chem. Biol. 2009, 13, 152–160. [Google Scholar] [CrossRef] [PubMed]
- Cartuche, L.; Reyes-Batlle, M.; Sifaoui, I.; Arberas-Jiménez, I.; Piñero, J.E.; Fernández, J.J.; Lorenzo-Morales, J.; Díaz-Marrero, A.R. Antiamoebic Activities of Indolocarbazole Metabolites Isolated from Streptomyces sanyensis Cultures. Mar. Drugs 2019, 17, 588. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Zhou, B.; Liu, H.; Huo, C.; Ding, W. One New Indolocarbazole Alkaloid from the Streptomyces sp. A22. Nat. Prod. Res. 2018, 32, 2583–2588. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Monger, A.; Wang, L.; Fu, P.; Piyachaturawat, P.; Chairoungdua, A.; Zhu, W. Precursor-Directed Generation of Indolocarbazoles with Topoisomerase IIα Inhibitory Activity. Mar. Drugs 2018, 16, 168. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.L.; Zhang, B.; Xue, W.W.; Li, W.; Xu, Z.F.; Shi, J.; Shen, Y.; Jiao, R.H.; Tan, R.X.; Ge, H.M. Discovery, Biosynthesis, and Heterologous Production of Loonamycin, a Potent Anticancer Indolocarbazole Alkaloid. Org. Lett. 2020, 22, 4665–4669. [Google Scholar] [CrossRef]
- Guan, J. Insulin-like Growth Factor-1 and Its Derivatives: Potential Pharmaceutical Application for Ischemic Brain Injury. Recent Pat. CNS Drug Discov. 2008, 3, 112–127. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Y.; Deng, J.; Jiang, H.; Zhuang, L.; Ye, W.; Ma, J.; Jiang, J.; Feng, L. Diketopiperazines Synthesis Gene in Shewanella Baltica and Roles of Diketopiperazines and Resveratrol in Quorum Sensing. J. Agric. Food Chem. 2019, 67, 12013–12025. [Google Scholar] [CrossRef]
- Turkez, H.; Cacciatore, I.; Arslan, M.E.; Fornasari, E.; Marinelli, L.; di Stefano, A.; Mardinoglu, A. Histidyl-Proline Diketopiperazine Isomers as Multipotent Anti-Alzheimer Drug Candidates. Biomolecules 2020, 10, 737. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Hou, Y.; Yang, Q.; Li, X.; Wu, S. Structures and Biological Activities of Diketopiperazines from Marine Organisms: A Review. Mar. Drugs 2021, 19, 403. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Chen, X.; Li, W.; Lu, C.; Shen, Y. New Diketopiperazine Derivatives with Cytotoxicity from Nocardiopsis sp. YIM M13066. J. Antibiot. 2017, 70, 795–797. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chai, W.; Zhu, R.; Song, T.; Zhang, Z.; Lian, X.Y. Streptopyrazinones A−D, Rare Metabolites from Marine-Derived Streptomyces sp. ZZ446. Tetrahedron 2018, 74, 2100–2106. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, D.; Chen, M.; Zhang, Z.; Lian, X.-Y. A Rare Diketopiperazine Glycoside from Marine-Sourced Streptomyces sp. ZZ446. Nat. Prod. Res. 2018, 34, 1046–1050. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Cullum, R.; Machado, H.; Smith, A.J.; Yang, I.; Rodvold, J.J.; Fenical, W. Photopiperazines A−D, Photosensitive Interconverting Diketopiperazines with Significant and Selective Activity against U87 Glioblastoma Cells, from a Rare, Marine-Derived Actinomycete of the Family Streptomycetaceae. J. Nat. Prod. 2019, 82, 2262–2267. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Qin, L.; Lian, X.-Y.; Zhang, Z. New Antifungal Metabolites from the Mariana Trench Sediment-Associated Actinomycete Streptomyces sp. SY1965. Mar. Drugs 2020, 18, 385. [Google Scholar] [CrossRef] [PubMed]
- Song, M.-M.; Xie, Y.-H.; Chen, W.-H.; Hu, Y.-W.; Zhao, K.; Liu, Y.-H.; Huang, X.-L.; Liu, Q.-C.; Wang, J.-F. Diketopiperazine and Enterotoxin Analogues from the Mangrove Derived-Soil Streptomyces sp. SCSIO 41400 and Their Biological Evaluation. Nat. Prod. Res. 2020, 1–8. [Google Scholar] [CrossRef]
- Kim, M.C.; Winter, J.M.; Asolkar, R.N.; Boonlarppradab, C.; Cullum, R.; Fenical, W. Marinoterpins A−C: Rare Linear Merosesterterpenoids from Marine-Derived Actinomycete Bacteria of the Family Streptomycetaceae. J. Org. Chem. 2021, 86, 11140–11148. [Google Scholar] [CrossRef]
- Tang, Z.; Niu, S.; Liu, F.; Lao, K.; Miao, J.; Ji, J.; Wang, X.; Yan, M.; Zhang, L.; You, Q.; et al. Synthesis and Biological Evaluation of 2,3-Diaryl Isoquinolinone Derivatives as Anti-Breast Cancer Agents Targeting ERα and VEGFR-2. Bioorg. Med. Chem. Lett. 2014, 24, 2129–2133. [Google Scholar] [CrossRef]
- Eliwa, E.M.; Abdel-Razek, A.S.; Frese, M.; Wibberg, D.; Halawa, A.H.; El-Agrody, A.M.; Bedair, A.H.; Kalinowski, J.; Sewald, N.; Shaaban, M. New Bioactive Compounds from the Marine-Derived Actinomycete Nocardiopsis Lucentensis sp. ASMR2. Zeitschrift Für Naturforschung B 2017, 72, 351–360. [Google Scholar] [CrossRef]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Rückert, C.; Braig, S.; Zahler, S.; et al. New Natural Products Identified by Combined Genomics-Metabolomics Profiling of Marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef]
- Kimura, T.; Inahashi, Y.; Matsuo, H.; Suga, T.; Iwatsuki, M.; Shiomi, K.; Takahashi, Y.; Ōmura, S.; Nakashima, T. Pyrizomicin A and B: Structure and Bioactivity of New Thiazolyl Pyridines from Lechevalieria Aerocolonigenes K10-0216. J. Antibiot. 2018, 71, 606–608. [Google Scholar] [CrossRef] [PubMed]
- Chanana, S.; Thomas, C.S.; Braun, D.R.; Hou, Y.; Wyche, T.P.; Bugni, T.S. Natural Product Discovery Using Planes of Principal Component Analysis in R (PoPCAR). Metabolites 2017, 7, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Y.; Braun, D.R.; Michel, C.R.; Klassen, J.L.; Adnani, N.; Wyche, T.P.; Bugni, T.S. Microbial Strain Prioritization Using Metabolomics Tools for the Discovery of Natural Products. Anal. Chem. 2012, 84, 4277. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Takahashi, Y.; Ōmura, S. Search for New Compounds from Kitasato Microbial Library by Physicochemical Screening. Biochem. Pharmacol. 2017, 134, 42–55. [Google Scholar] [CrossRef]
- Reen, F.J.; Romano, S.; Dobson, A.D.W.; O’Gara, F. The Sound of Silence: Activating Silent Biosynthetic Gene Clusters in Marine Microorganisms. Mar. Drugs 2015, 13, 4754–4783. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.-W.; Lin, L.-G.; Ye, W.-C. Techniques for Extraction and Isolation of Natural Products: A Comprehensive Review. Chin. Med. 2018, 13, 20. [Google Scholar] [CrossRef] [Green Version]
- Subramani, R.; Aalbersberg, W. Marine Actinomycetes: An Ongoing Source of Novel Bioactive Metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Rutledge, P.J.; Challis, G.L. Discovery of Microbial Natural Products by Activation of Silent Biosynthetic Gene Clusters. Nat. Rev. Microbiol. 2015, 13, 509–523. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Hughes, D.E.; Epstein, S.; Jones, M.; et al. A New Antibiotic Kills Pathogens without Detectable Resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Albarano, L.; Esposito, R.; Ruocco, N.; Costantini, M. Genome Mining as New Challenge in Natural Products Discovery. Mar. Drugs 2020, 18, 199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, N.; Kim, W.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.K. Thirty Complete Streptomyces Genome Sequences for Mining Novel Secondary Metabolite Biosynthetic Gene Clusters. Sci. Data 2020, 7, 55. [Google Scholar] [CrossRef]
- Minoche, A.E.; Dohm, J.C.; Himmelbauer, H. Evaluation of Genomic High-Throughput Sequencing Data Generated on Illumina HiSeq and Genome Analyzer Systems. Genome Biol. 2011, 12, R112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weirather, J.L.; de Cesare, M.; Wang, Y.; Piazza, P.; Sebastiano, V.; Wang, X.-J.; Buck, D.; Au, K.F. Comprehensive Comparison of Pacific Biosciences and Oxford Nanopore Technologies and Their Applications to Transcriptome Analysis. F1000Research 2017, 6, 100. [Google Scholar] [CrossRef] [PubMed]
- Baltz, R.H. Molecular Beacons to Identify Gifted Microbes for Genome Mining. J. Antibiot. 2017, 70, 639–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Completing Bacterial Genome Assemblies with Multiplex MinION Sequencing. Microb. Genom. 2017, 3, e000132. [Google Scholar] [CrossRef]
- Salzberg, S.L. Next-Generation Genome Annotation: We Still Struggle to Get It Right. Genome Biol. 2019, 20, 92. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Holt, K.E. Benchmarking of Long-Read Assemblers for Prokaryote Whole Genome Sequencing. F1000Research 2019, 8, 2138. [Google Scholar] [CrossRef] [Green Version]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A Parallel Assembler for Short Read Sequence Data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [Green Version]
- Zimin, A.V.; Marçais, G.; Puiu, D.; Roberts, M.; Salzberg, S.L.; Yorke, J.A. The MaSuRCA Genome Assembler. Bioinformatics 2013, 29, 2669–2677. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; Dicuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, C.; Kim, K.; Cui, L.; Liu, X. Genome Annotation of Disease-Causing Microorganisms. Brief. Bioinform. 2021, 22, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded Prediction of Natural Product Chemical Structures from Microbial Genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B.-K. Mini Review: Genome Mining Approaches for the Identification of Secondary Metabolite Biosynthetic Gene Clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef]
- Cimermancic, P.; Medema, M.H.; Claesen, J.; Kurita, K.; Brown, L.C.W.; Mavrommatis, K.; Pati, A.; Godfrey, P.A.; Koehrsen, M.; Clardy, J.; et al. Insights into Secondary Metabolism from a Global Analysis of Prokaryotic Biosynthetic Gene Clusters. Cell 2014, 158, 412–421. [Google Scholar] [CrossRef] [Green Version]
- Hannigan, G.D.; Prihoda, D.; Palicka, A.; Soukup, J.; Klempir, O.; Rampula, L.; Durcak, J.; Wurst, M.; Kotowski, J.; Chang, D.; et al. A Deep Learning Genome-Mining Strategy for Biosynthetic Gene Cluster Prediction. Nucleic Acids Res. 2019, 47, e110. [Google Scholar] [CrossRef]
- Chavali, A.K.; Rhee, S.Y. Bioinformatics Tools for the Identification of Gene Clusters That Biosynthesize Specialized Metabolites. Brief. Bioinform. 2018, 19, 1022–1034. [Google Scholar] [CrossRef]
- Chu, L.; Huang, J.; Muhammad, M.; Deng, Z.; Gao, J. Genome Mining as a Biotechnological Tool for the Discovery of Novel Marine Natural Products. Crit. Rev. Biotechnol. 2020, 40, 571–589. [Google Scholar] [CrossRef]
- Crüsemann, M. Coupling Mass Spectral and Genomic Information to Improve Bacterial Natural Product Discovery Workflows. Mar. Drugs 2021, 19, 142. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Molloy, B.; Braña, A.F.; Zabala, D.; Olano, C.; Cortés, J.; Morís, F.; Salas, J.A.; Méndez, C. Identification by Genome Mining of a Type I Polyketide Gene Cluster from Streptomyces argillaceus Involved in the Biosynthesis of Pyridine and Piperidine Alkaloids Argimycins P. Front. Microbiol. 2017, 8, 194. [Google Scholar] [CrossRef] [Green Version]
- Sun, C.; Yang, Z.; Zhang, C.; Liu, Z.; He, J.; Liu, Q.; Zhang, T.; Ju, J.; Ma, J. Genome Mining of Streptomyces atratus SCSIO ZH16: Discovery of Atratumycin and Identification of Its Biosynthetic Gene Cluster. Org. Lett. 2019, 21, 1453–1457. [Google Scholar] [CrossRef]
- Lee, N.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Synthetic Biology Tools for Novel Secondary Metabolite Discovery in Streptomyces. J. Microbiol. Biotechnol. 2019, 29, 667–686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Xiao, L.; Zhou, Y.; Deng, K.; Tan, G.; Han, Y.; Liu, X.; Deng, Z.; Liu, T. Development of Streptomyces sp. FR-008 as an Emerging Chassis. Synth. Syst. Biotechnol. 2016, 1, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Begley, T.; Liu, H. Comprehensive Natural Products III: Chemistry Biology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2020; ISBN 9780081026915. [Google Scholar]
- Li, L.; Maclntyre, L.W.; Brady, S.F. Refactoring Biosynthetic Gene Clusters for Heterologous Production of Microbial Natural Products. Curr. Opin. Biotechnol. 2021, 69, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhao, Y.; Huang, C.; Luo, Y. Recent Advances in Silent Gene Cluster Activation in Streptomyces. Front. Bioeng. Biotechnol. 2021, 9, 632230. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, M.; Du, G.; Chen, J. Improving the Active Expression of Transglutaminase in Streptomyces lividans by Promoter Engineering and Codon Optimization. BMC Biotechnol. 2016, 16, 75. [Google Scholar] [CrossRef] [Green Version]
- Panchal, V.; Brenk, R. Riboswitches as Drug Targets for Antibiotics. Antibiotics 2021, 10, 45. [Google Scholar] [CrossRef]














Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Rop, A.-S.; Rombaut, J.; Willems, T.; De Graeve, M.; Vanhaecke, L.; Hulpiau, P.; De Maeseneire, S.L.; De Mol, M.L.; Soetaert, W.K. Novel Alkaloids from Marine Actinobacteria: Discovery and Characterization. Mar. Drugs 2022, 20, 6. https://doi.org/10.3390/md20010006
De Rop A-S, Rombaut J, Willems T, De Graeve M, Vanhaecke L, Hulpiau P, De Maeseneire SL, De Mol ML, Soetaert WK. Novel Alkaloids from Marine Actinobacteria: Discovery and Characterization. Marine Drugs. 2022; 20(1):6. https://doi.org/10.3390/md20010006
Chicago/Turabian StyleDe Rop, Anne-Sofie, Jeltien Rombaut, Thomas Willems, Marilyn De Graeve, Lynn Vanhaecke, Paco Hulpiau, Sofie L. De Maeseneire, Maarten L. De Mol, and Wim K. Soetaert. 2022. "Novel Alkaloids from Marine Actinobacteria: Discovery and Characterization" Marine Drugs 20, no. 1: 6. https://doi.org/10.3390/md20010006