Characterization of Bioactivities and Biosynthesis of Angucycline/Angucyclinone Derivatives Derived from Gephyromycinifex aptenodytis gen. nov., sp. nov.
Abstract
:1. Introduction
2. Results and Discussion
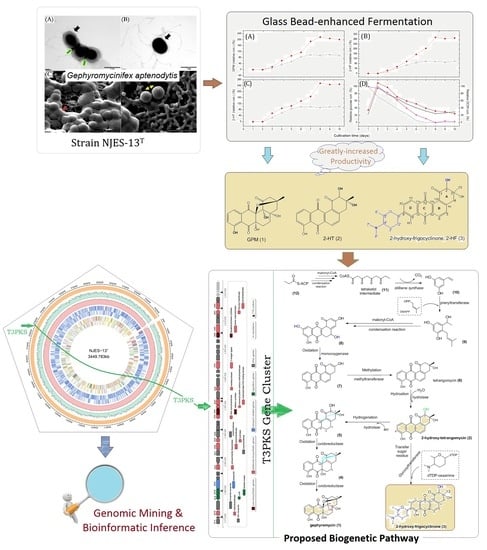
2.1. Fermentation Based on Glass Bead-Enhanced Cultivation of Strain NJES-13T
2.2. New Angucyclinone Derivative Metabolite Characterization
2.3. Bioactivity Assays of Three AD Metabolites
2.4. Biosynthetic Gene Analysis
2.5. Proposed Biogenetic Pathway for Three AD Metabolites
3. Materials and Methods
3.1. Bacterial Culture
3.2. Glass Bead-Enhanced Fermentation
3.3. Bacterial Metabolite Isolation
3.4. Chemical Structure Characterization
3.5. Quantitative Analysis of Metabolites
3.6. Glucose and Dry Cell Weight Concentration Measurements
3.7. Bioactivity Evaluations
3.8. Biosynthetic Pathway Analysis of ADs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Subramani, R.; Sipkema, D. Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 7, 249. [Google Scholar] [CrossRef] [Green Version]
- Atencio, L.A.; Boya, P.C.A.; Martin, H.C.; Mejía, L.C.; Dorrestein, P.C.; Gutiérrez, M. Genome Mining, Microbial Interactions, and Molecular Networking Reveals New Dibromoalterochromides from Strains of Pseudoalteromonas of Coiba National Park-Panama. Mar. Drugs 2020, 18, 456. [Google Scholar] [CrossRef]
- Lee, N.; Hwang, S.; Kim, W.; Lee, Y.; Kim, J.H.; Cho, S.; Kim, H.U.; Yoon, Y.J.; Oh, M.K.; Palsson, B.O.; et al. Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in Streptomyces genomes. Nat. Prod. Rep. 2021, 38, 1330–1361. [Google Scholar] [CrossRef]
- Chen, J.; Xu, L.; Zhou, Y.; Han, B. Natural Products from Actinomycetes Associated with Marine Organisms. Mar. Drugs 2021, 19, 629. [Google Scholar] [CrossRef]
- Kharel, M.K.; Pahari, P.; Shepherd, M.D.; Tibrewal, N.; Nybo, S.E.; Shaaban, K.A.; Rohr, J. Angucyclines: Biosynthesis, mode-of-action, new natural products, and synthesis. Nat. Prod. Rep. 2012, 29, 264–325. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, H.P. Abyssomicins-A 20-Year Retrospective View. Mar. Drugs 2021, 19, 299. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, S.A.M.; Elias, N.; Farag, M.A.; Chen, L.; Saeed, A.; Hegazy, M.F.; Moustafa, M.S.; Abd El-Wahed, A.; Al-Mousawi, S.M.; Musharraf, S.G.; et al. Marine Natural Products: A Source of Novel Anticancer Drugs. Mar. Drugs 2019, 17, 491. [Google Scholar] [CrossRef] [Green Version]
- Gerhard, B.; Gerhard, L.; Kadja, M.; Andreas, H.; Tobias, A.M.G.; Anke, D.; Alan, T.B.; James, E.M.S.; Niko, K.; Werner, E.G.M. Gephyromycin, the first bridged angucyclinone, from Streptomyces griseus strain NTK 14. Phytochemistry 2005, 66, 1366–1373. [Google Scholar]
- Kern, D.L.; Schaumberg, J.P.; Hokanson, G.C.; French, J.C. PD 116,779, a new antitumor antibiotic of the benz[a]anthraquinone class. J. Antibiot. 1986, 39, 469–470. [Google Scholar] [CrossRef] [Green Version]
- Ding, W.J.; Ji, Y.Y.; Jiang, Y.J.; Ying, W.J.; Fang, Z.Y.; Gao, T.T. Gephyromycin C, a novel small-molecule inhibitor of heat shock protein Hsp90, induces G2/M cell cycle arrest and apoptosis in PC3 cells in vitro. Biochem. Biophys. Res. Commun. 2020, 531, 377–382. [Google Scholar] [CrossRef]
- Gao, H.M.; Xie, P.F.; Zhang, X.L.; Yang, Q. Isolation, phylogenetic and gephyromycin metabolites characterization of new exopolysaccharides-bearing antarctic actinobacterium from faeces of emperor penguin. Mar. Drugs 2021, 19, 458. [Google Scholar] [CrossRef]
- Zhu, W.Z.; Ge, Y.M.; Gao, H.M.; Dai, J.; Zhang, X.L.; Yang, Q. Gephyromycinifex aptenodytis gen. nov., sp. nov., isolated from gut of Antarctic emperor penguin. Antonie Van Leeuwenhoek 2021, 114, 2003–2017. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Liu, B.; Wang, H.; Yang, S.; Zhang, H.; Wang, Y.; Ji, N.; Qin, S.; Laatsch, H. Kiamycin, a unique cytotoxic angucyclinone derivative from a marine Streptomyces sp. Mar. Drugs 2012, 10, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.; Gan, L.S.; Ding, W.J.; Chen, Z.; Ma, Z.J. Cytotoxic gephyromycins from the Streptomyces sp. SS13I. Tetrahedron Lett. 2017, 58, 3747–3750. [Google Scholar] [CrossRef]
- Chang, Y.; Xing, L.; Sun, C.; Liang, S.; Liu, T.; Zhang, X.; Zhu, T.; Pfeifer, B.A.; Che, Q.; Zhang, G.; et al. Monacycliones G-K and ent-Gephyromycin A, Angucycline Derivatives from the Marine-Derived Streptomyces sp. HDN15129. J. Nat. Prod. 2020, 83, 2749–2755. [Google Scholar] [CrossRef]
- Duan, Y.; Jiang, Z.; Wu, Z.; Sheng, Z.; Yang, X.; Sun, J.; Zhang, X.; Yang, Q.; Yu, X.; Yan, J. Limnobacter alexandrii sp. nov., a thiosulfate-oxidizing, heterotrophic and EPS-bearing Burkholderiaceae isolated from cultivable phycosphere microbiota of toxic Alexandrium catenella LZT09. Antonie Van Leeuwenhoek 2020, 113, 1689–1698. [Google Scholar] [CrossRef]
- Yang, Q.; Ge, Y.M.; Iqbal, N.M.; Yang, X.; Zhang, X.L. Sulfitobacter alexandrii sp. nov., a new microalgae growth-promoting bacterium with exopolysaccharides bioflocculanting potential isolated from marine phycosphere. Antonie Van Leeuwenhoek 2021, 114, 1091–1106. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Qi, M.; Li, Q.H.; Cui, Z.D.; Yang, Q. Maricaulis alexandrii sp. nov., a novel active bioflocculants-bearing and dimorphic prosthecate bacterium isolated from marine phycosphere. Antonie Van Leeuwenhoek 2021, 114, 1195–1203. [Google Scholar] [CrossRef]
- Van der Heul, H.U.; Bilyk, B.L.; McDowall, K.J.; Seipke, R.F.; van Wezel, G.P. Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Chater, K.F.; Chandra, G.; Niu, G.; Tan, H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol. Mol. Biol. Rev. 2013, 77, 112–143. [Google Scholar] [CrossRef] [Green Version]
- Böl, M.; Schrinner, K.; Tesche, S.; Krull, R. Challenges of influencing cellular morphology by morphology engineering techniques and mechanical induced stress on filamentous pellet systems—A critical review. Eng. Life Sci. 2020, 21, 51–67. [Google Scholar] [CrossRef]
- Fanning, S.; Hall, L.J.; Cronin, M.; Zomer, A.; MacSharry, J.; Goulding, D.; Motherway, M.O.; Shanahan, F.; Nally, K.; Dougan, G.; et al. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc. Natl. Acad. Sci. USA 2012, 109, 2108–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaup, B.A.; Ehrich, K.; Pescheck, M.; Schrader, J. Microparticle-enhanced cultivation of filamentous microorganisms: Increased chloroperoxidase formation by Caldariomyces fumago as an example. Biotechnol. Bioeng. 2008, 99, 491–498. [Google Scholar] [CrossRef]
- Lin, P.J.; Scholz, A.; Krull, R. Effect of volumetric power input by aeration and agitation on pellet morphology and product formation of Aspergillus niger. Biochem. Eng. J. 2010, 49, 213–220. [Google Scholar] [CrossRef]
- Mo, J.; Ye, J.; Chen, H.; Hou, B.; Wu, H.; Zhang, H. Cloning and identification of the Frigocyclinone biosynthetic gene cluster from Streptomyces griseus strain NTK 97. Biosci. Biotechnol. Biochem. 2019, 83, 2082–2089. [Google Scholar] [CrossRef]
- Balachandran, C.; Arun, Y.; Duraipandiyan, V.; Ignacimuthu, S.; Balakrishna, K.; Al-Dhabi, N.A. Antimicrobial and cytotoxicity properties of 2,3-dihydroxy-9,10-anthraquinone isolated from Streptomyces galbus (ERINLG-127). Appl. Biochem. Biotechnol. 2014, 172, 3513–3528. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 12, gkab335. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, X.A.; Jiang, Z.W.; Yang, X.; Zhang, X.L.; Yang, Q. Combined characterization of a new member of Marivita cryptomonadis, strain LZ-15-2 isolated from cultivable phycosphere microbiota of toxic HAB dinoflagellate Alexandrium catenella LZT09. Braz. J. Microbiol. 2021, 52, 739–748. [Google Scholar] [CrossRef]
- Zhang, X.L.; Li, G.X.; Ge, Y.M.; Iqbal, M.N.; Yang, X.; Cui, Z.D.; Yang, Q. Sphingopyxis microcytisis sp. nov., a novel bioactive exopolysaccharides-bearing Sphingomonadaceae isolated from the Microcytis phycosphere. Antonie Van Leeuwenhoek 2021, 114, 845–857. [Google Scholar] [CrossRef]
- Deng, Z.; Liu, J.; Li, T.; Li, H.; Liu, Z.; Dong, Y.; Li, W. An Unusual type III Polyketide Synthase System Involved in Cinnamoyl Lipid Biosynthesis. Angew. Chem. Int. Ed. Engl. 2021, 60, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, Y.; Ishikawa, J.; Hara, H.; Suzuki, H.; Ikenoya, M.; Ikeda, H.; Yamashita, A.; Hattori, M.; Horinouchi, S. Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 2008, 190, 4050–4060. [Google Scholar] [CrossRef] [Green Version]
- Baerson, S.R.; Schröder, J.; Cook, D.; Rimando, A.M.; Pan, Z.; Dayan, F.E.; Noonan, B.P.; Duke, S.O. Alkylresorcinol biosynthesis in plants: New insights from an ancient enzyme family? Plant Signal. Behav. 2010, 5, 1286–1289. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, R.; Chen, X.; Sun, X.; Yan, Y.; Shen, X.; Yuan, Q. Biosynthesis of aromatic polyketides in microorganisms using type II polyketide synthases. Microb. Cell Fact. 2020, 19, 110. [Google Scholar] [CrossRef]
- Härle, J.; Günther, S.; Lauinger, B.; Weber, M.; Kammerer, B.; Zechel, D.L.; Luzhetskyy, A.; Bechthold, A. Rational design of an aryl-C-glycoside catalyst from a natural product O-glycosyltransferase. Chem. Biol. 2011, 18, 520–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.; Feng, Q.; Zhang, B.P.; Gao, J.J.; Sheng, Z.; Xue, Q.P.; Zhang, X.L. Marinobacter alexandrii sp. nov., a novel yellow-pigmented and algae growth-promoting bacterium isolated from marine phycosphere microbiota. Antonie Van Leeuwenhoek 2021, 114, 709–718. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, X.L.; Li, L.; Zhang, R.N.; Feng, L.J.; Mu, J. Ponticoccus alexandrii sp. nov., a novel bacterium isolated from the marine toxigenic dinoflagellate Alexandrium minutum. Antonie Van Leeuwenhoek 2018, 11, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Houdkova, M.; Albarico, G.; Doskocil, I.; Tauchen, J.; Urbanova, K.; Tulin, E.E.; Kokoska, L. Vapors of Volatile Plant-Derived Products Significantly Affect the Results of Antimicrobial, Antioxidative and Cytotoxicity Microplate-Based Assays. Molecules 2020, 25, 6004. [Google Scholar] [CrossRef]
- Liu, J.Z.; Yang, J.S.; Ge, Y.M.; Yang, Q.; Sun, J.Y.; Yu, X. Acute effects of CH3NH3PbI3 perovskite on Scenedesmus obliquus and Daphnia magana in aquatic environment. Ecotoxicol. Environ. Saf. 2021, 208, 111677. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the MTT Assay. Cold Spring Harb. Protoc. 2018, 2018, pdb-prot095505. [Google Scholar] [CrossRef]
- Yang, Q.; Jiang, Z.; Zhou, X.; Xie, Z.; Wang, Y.; Wang, D.; Feng, L.; Yang, G.; Ge, Y.; Zhang, X. Saccharospirillum alexandrii sp. nov., isolated from the toxigenic marine dinoflagellate Alexandrium catenella LZT09. Int. J. Syst. Evol. Microbiol. 2020, 70, 820–826. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, Z.; Zhou, X.; Zhang, R.; Wu, Y.; Lou, L.; Ma, Z.; Wang, D.; Ge, Y.; Zhang, X.; et al. Nioella ostreopsis sp. nov., isolated from toxic dinoflagellate, Ostreopsis lenticularis. Int. J. Syst. Evol. Microbiol. 2020, 70, 759–765. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Jiang, Z.; Zhou, X.; Zhang, R.; Xie, Z.; Zhang, S.; Wu, Y.; Ge, Y.; Zhang, X. Haliea alexandrii sp. nov., isolated from phycosphere microbiota of the toxin-producing dinoflagellate Alexandrium catenella. Int. J. Syst. Evol. Microbiol. 2020, 70, 1133–1138. [Google Scholar] [CrossRef]
- Yang, X.; Jiang, Z.W.; Zhang, J.; Zhou, X.; Zhang, X.L.; Wang, L.; Yu, T.; Wang, Z.; Bei, J.; Dong, B. Mesorhizobium alexandrii sp. nov., isolated from phycosphere microbiota of PSTs-producing marine dinoflagellate Alexandrium minutum amtk4. Antonie Van Leeuwenhoek 2020, 113, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, L.; Redou, V.; Cochereau, B.; Delage, L.; Hymery, N.; Poirier, E.; Le Meur, C.; Le Foch, G.; Cladiere, L.; Mehiri, M.; et al. Identification and Characterization of a New type III Polyketide Synthase from a Marine Yeast, Naganishia uzbekistanensis. Mar. Drugs 2020, 18, 637. [Google Scholar] [CrossRef]
- Dai, J.; Li, K.; Song, N.; Yao, W.; Xia, H.; Yang, Q.; Zhang, X.; Li, X.; Wang, Z.; Yao, L.; et al. Zygosaccharomyces rouxii, an Aromatic Yeast Isolated From Chili Sauce, Is Able to Biosynthesize 2-Phenylethanol via the Shikimate or Ehrlich Pathways. Front. Microbiol. 2020, 11, 597454. [Google Scholar] [CrossRef]
- Prichula, J.; Primon-Barros, M.; Luz, R.C.Z.; Castro, Í.M.S.; Paim, T.G.S.; Tavares, M.; Ligabue-Braun, R.; d’Azevedo, P.A.; Frazzon, J.; Frazzon, A.P.G.; et al. Genome Mining for Antimicrobial Compounds in Wild Marine Animals-Associated Enterococci. Mar. Drugs 2021, 19, 328. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, H.; Bowler, C.; Xing, X.; Bulone, V.; Shao, Z.; Duan, D. Full-Length Transcriptome of Thalassiosira weissflogii as a Reference Resource and Mining of Chitin-Related Genes. Mar. Drugs 2021, 19, 392. [Google Scholar] [CrossRef]






| Position | δH | δC | Position | δH | δC |
|---|---|---|---|---|---|
| 1 | — | 192.5, Cq | 12 | — | 172.1, Cq |
| 2 | 4.90 | 75.2, CH | 12a | — | 138.2, Cq |
| 3 | — | 73.2, Cq | 12b | — | 139.1, Cq |
| 4 | 2.63 | 42.2, CH2 | 13 | 1.32 (s) | 20.3, CH3 |
| 4a | — | 147.2, Cq | 1’ | 4.92ax (m) | 78.7, CH |
| 5 | 7.57 | 132.6, CH | 2’ | 2.05eq/1.45ax (m) | 30.7, CH2 |
| 6 | 7.92 (d, 8.0) | 129.9, CH | 3’ | 1.84eq/1.58ax (m) | 20.2, CH2 |
| 6a | — | 132.5, Cq | 4’ | 2.25ax (m) | 72.4, CH |
| 7 | — | 181.0, Cq | 5’ | 3.98eq (m) | 78.5, CH |
| 7a | — | 118.5, Cq | 6’ | 1.21 (d, 6.8) | 10.7, CH3 |
| 8 | — | 159.9, Cq | 7’, 8’ | 2.20 (s) | 41.8, CH3 |
| 9 | — | 138.4, Cq | 2-OH | 6.04 (s) | — |
| 10 | 7.40 | 132.4, CH | 3-OH | 4.95 (br) | — |
| 11 | 7.43 | 120.7, CH | 8-OH | 12.10 (s, br) | — |
| 11a | — | 136.2, Cq |
| Tested Compounds | Antibacterial Activity (MIC, µg/mL) | Cytotoxic Activity (IC50, µM) | ||||
|---|---|---|---|---|---|---|
| B. subtilis | S. aureus | C. albicans | HL-60 | Bel-7402 | A549 | |
| Gephyromycin (GPM) | >100 | >100 | >100 | 133.2 | 108.7 | 154.3 |
| 2-Hydroxy-tetrangomycin (2-HT) | 27.2 | 14.1 | 15.6 | 25.8 | 35.6 | 14.1 |
| 2-Hydroxy-frigocyclinone (2-HF) | 15.4 | 5.7 | 8.5 | 8.4 | 4.2 | 5.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, W.-Z.; Wang, S.-H.; Gao, H.-M.; Ge, Y.-M.; Dai, J.; Zhang, X.-L.; Yang, Q. Characterization of Bioactivities and Biosynthesis of Angucycline/Angucyclinone Derivatives Derived from Gephyromycinifex aptenodytis gen. nov., sp. nov. Mar. Drugs 2022, 20, 34. https://doi.org/10.3390/md20010034
Zhu W-Z, Wang S-H, Gao H-M, Ge Y-M, Dai J, Zhang X-L, Yang Q. Characterization of Bioactivities and Biosynthesis of Angucycline/Angucyclinone Derivatives Derived from Gephyromycinifex aptenodytis gen. nov., sp. nov. Marine Drugs. 2022; 20(1):34. https://doi.org/10.3390/md20010034
Chicago/Turabian StyleZhu, Wen-Zhuo, Shu-Heng Wang, Hui-Min Gao, Ya-Ming Ge, Jun Dai, Xiao-Ling Zhang, and Qiao Yang. 2022. "Characterization of Bioactivities and Biosynthesis of Angucycline/Angucyclinone Derivatives Derived from Gephyromycinifex aptenodytis gen. nov., sp. nov." Marine Drugs 20, no. 1: 34. https://doi.org/10.3390/md20010034