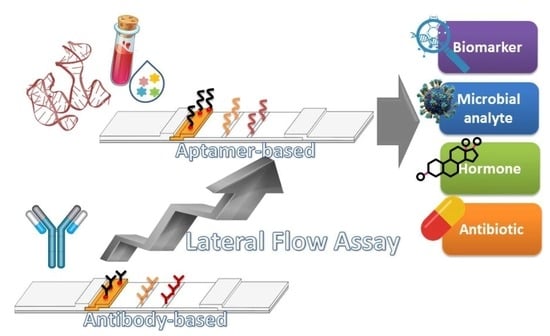
Aptamer-Based Lateral Flow Assays: Current Trends in Clinical Diagnostic Rapid Tests
Abstract
:1. Introduction
2. Design and Principle of Lateral Flow Assays


3. Replacing Aptamers with Antibodies in LFAs
4. Clinical Applications of Aptamer-Based LFAs
4.1. Detection of Biomarkers



4.2. Detection of Microbial Analytes
4.3. Detection of Hormones
4.4. Detection of Antibiotics
5. Evaluation of Sensitivity and Specificity of Aptamer-Based LFAs in Comparison with Antibody-Based LFAs
6. Current and Future Challenges of LFAs
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Koczula, K.M.; Gallotta, A. Lateral flow assays. Essays Biochem. 2016, 60, 111–120. [Google Scholar] [PubMed]
- Jauset-Rubio, M.; El-Shahawi, M.S.; Bashammakh, A.S.; Alyoubi, A.O.; Ciara, K. Advances in aptamers-based lateral flow assays. TrAC Trends Anal. Chem. 2017, 97, 385–398. [Google Scholar] [CrossRef]
- Majdinasab, M.; Zareian, M.; Zhang, Q.; Li, P. Development of a new format of competitive immunochromatographic assay using secondary antibody–europium nanoparticle conjugates for ultrasensitive and quantitative determination of ochratoxin A. Food Chem. 2019, 275, 721–729. [Google Scholar] [CrossRef] [PubMed]
- Xiao, X.; Hu, S.; Lai, X.; Peng, J.; Lai, W. Developmental trend of immunoassays for monitoring hazards in food samples: A review. Trends Food Sci. Technol. 2021, 111, 68–88. [Google Scholar] [CrossRef]
- Sadeghi, P.; Sohrabi, H.; Hejazi, M.; Jahanban-Esfahlan, A.; Baradaran, B.; Tohidast, M.; Majidi, M.R.; Mokhtarzadeh, A.; Tavangar, S.M.; de la Guardia, M. Lateral flow assays (LFA) as an alternative medical diagnosis method for detection of virus species: The intertwine of nanotechnology with sensing strategies. TrAC Trends Anal. Chem. 2021, 145, 116460. [Google Scholar] [CrossRef]
- Bahadır, E.B.; Sezgintürk, M.K. Lateral flow assays: Principles, designs and labels. TrAC Trends Anal. Chem. 2016, 82, 286–306. [Google Scholar] [CrossRef]
- Majdinasab, M.; Sheikh-Zeinoddin, M.; Soleimanian-Zad, S.; Li, P.; Zhang, Q.; Li, X.; Tang, X. Ultrasensitive and quantitative gold nanoparticle-based immunochromatographic assay for detection of ochratoxin A in agro-products. J. Chromatogr. B 2015, 974, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Mak, W.C.; Beni, V.; Turner, A.P. Lateral-flow technology: From visual to instrumental. TrAC Trends Anal. Chem. 2016, 79, 297–305. [Google Scholar] [CrossRef]
- Sajid, M.; Kawde, A.-N.; Daud, M. Designs, formats and applications of lateral flow assay: A literature review. J. Saudi Chem. Soc. 2015, 19, 689–705. [Google Scholar] [CrossRef] [Green Version]
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral flow (immuno) assay: Its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. [Google Scholar] [CrossRef] [Green Version]
- Raysyan, A.; Galvidis, I.A.; Schneider, R.J.; Eremin, S.A.; Burkin, M.A. Development of a latex particles-based lateral flow immunoassay for group determination of macrolide antibiotics in breast milk. J. Pharm. Biomed. Anal. 2020, 189, 113450. [Google Scholar] [CrossRef]
- Sharma, R.; Verma, A.; Shinde, N.; Mann, B.; Gandhi, K.; Wichers, J.H.; van Amerongen, A. Adulteration of cow’s milk with buffalo’s milk detected by an on-site carbon nanoparticles-based lateral flow immunoassay. Food Chem. 2021, 351, 129311. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.-H.; Huang, W.-C.; Chang, S.-C.; Kao, C.-H.; Shyu, R.-H. Colloidal silver-based lateral flow immunoassay for rapid detection of melamine in milk and animal feed. Mater. Chem. Phys. 2019, 231, 121–130. [Google Scholar] [CrossRef]
- Mao, X.; Wang, W.; Du, T.E. Dry-reagent nucleic acid biosensor based on blue dye doped latex beads and lateral flow strip. Talanta 2013, 114, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.W.; Kim, K.R.; Chun, H.J.; Jeong, K.Y.; Hong, D.-K.; Lee, K.-N.; Yoon, H.C. Time-resolved fluorescence resonance energy transfer-based lateral flow immunoassay using a raspberry-type europium particle and a single membrane for the detection of cardiac troponin I. Biosens. Bioelectron. 2020, 163, 112284. [Google Scholar] [CrossRef]
- Shang, Y.; Cai, S.; Ye, Q.; Wu, Q.; Shao, Y.; Qu, X.; Xiang, X.; Zhou, B.; Ding, Y.; Chen, M. Quantum dot nanobeads-labelled lateral flow immunoassay strip for rapid and sensitive detection of Salmonella Typhimurium based on strand displacement loop-mediated isothermal amplification. Engineering 2021. [Google Scholar] [CrossRef]
- Thiruvengadam, M.; Venkidasamy, B.; Subramanian, U. Up-converting phosphor technology-based lateral flow assay for quantitative detection of β-hydroxybutyrate in biological samples. Anal. Biochem. 2020, 591, 113546. [Google Scholar] [CrossRef]
- Wang, Z.; Zhi, D.; Zhao, Y.; Zhang, H.; Wang, X.; Ru, Y.; Li, H. Lateral flow test strip based on colloidal selenium immunoassay for rapid detection of melamine in milk, milk powder, and animal feed. Int. J. Nanomed. 2014, 9, 1699. [Google Scholar] [CrossRef] [Green Version]
- Moyano, A.; Serrano-Pertierra, E.; Salvador, M.; Martínez-García, J.C.; Rivas, M.; Blanco-López, M.C. Magnetic lateral flow immunoassays. Diagnostics 2020, 10, 288. [Google Scholar] [CrossRef]
- Pan, Q.; Luo, F.; Liu, M.; Zhang, X.-L. Oligonucleotide aptamers: Promising and powerful diagnostic and therapeutic tools for infectious diseases. J. Infect. 2018, 77, 83–98. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhao, J.; Li, J.; Liao, X.; Chen, F. Advances of aptamers screened by Cell-SELEX in selection procedure, cancer diagnostics and therapeutics. Anal. Biochem. 2020, 598, 113620. [Google Scholar] [CrossRef] [PubMed]
- Farzin, L.; Shamsipur, M.; Moassesi, M.E.; Sheibani, S. Clinical aspects of radiolabeled aptamers in diagnostic nuclear medicine: A new class of targeted radiopharmaceuticals. Bioorganic Med. Chem. 2019, 27, 2282–2291. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Yang, S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Zhu, L.; Hao, H.; Zhang, Z.; Ding, C.; Zhang, G.; Bi, J.; Yan, S.; Liu, G.; Hou, H. A novel photoelectrochemical aptamer sensor based on rare-earth doped Bi2WO6 and Ag2S for the rapid detection of Vibrio parahaemolyticus. Microchem. J. 2021, 165, 106132. [Google Scholar] [CrossRef]
- Nxele, S.R.; Nkhahle, R.; Nyokong, T. The composites of asymmetric Co phthalocyanines-graphitic carbon nitride quantum dots-aptamer as specific electrochemical sensors for the detection of prostate specific antigen: Effects of ring substituents. J. Electroanal. Chem. 2021, 900, 115730. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Liu, Y.; Chu, T.; Li, Y.; Dai, C.; Yang, Y.; Zhang, Y. Surface plasma enhanced fluorescence combined aptamer sensor based on silica modified silver nanoparticles for signal amplification detection of cholic acid. Microchem. J. 2021, 168, 106524. [Google Scholar] [CrossRef]
- Majdinasab, M.; Daneshi, M.; Marty, J.L. Recent developments in non-enzymatic (bio) sensors for detection of pesticide residues: Focusing on antibody, aptamer and molecularly imprinted polymer. Talanta 2021, 232, 122397. [Google Scholar] [CrossRef]
- Xu, H.; Mao, X.; Zeng, Q.; Wang, S.; Kawde, A.-N.; Liu, G. Aptamer-functionalized gold nanoparticles as probes in a dry-reagent strip biosensor for protein analysis. Anal. Chem. 2009, 81, 669–675. [Google Scholar] [CrossRef]
- Wang, L.; Ma, W.; Chen, W.; Liu, L.; Ma, W.; Zhu, Y.; Xu, L.; Kuang, H.; Xu, C. An aptamer-based chromatographic strip assay for sensitive toxin semi-quantitative detection. Biosens. Bioelectron. 2011, 26, 3059–3062. [Google Scholar] [CrossRef]
- Pai, N.P.; Vadnais, C.; Denkinger, C.; Engel, N.; Pai, M. Point-of-care testing for infectious diseases: Diversity, complexity, and barriers in low-and middle-income countries. PLoS Med. 2012, 9, e1001306. [Google Scholar] [CrossRef] [Green Version]
- Soh, J.H.; Chan, H.-M.; Ying, J.Y. Strategies for developing sensitive and specific nanoparticle-based lateral flow assays as point-of-care diagnostic device. Nano Today 2020, 30, 100831. [Google Scholar] [CrossRef]
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten years of lateral flow immunoassay technique applications: Trends, challenges and future perspectives. Sensors 2021, 21, 5185. [Google Scholar] [CrossRef] [PubMed]
- Market, F.C. Lateral flow immunoassay systems: Evolution from the current state of the art to the next generation of highly sensitive, quantitative rapid assays. Immunoass. Handb. 2013, 89, 89–107. [Google Scholar]
- Wang, K.K.; Yang, Z.; Zhu, T.; Shi, Y.; Rubenstein, R.; Tyndall, J.A.; Manley, G.T. An update on diagnostic and prognostic biomarkers for traumatic brain injury. Expert Rev. Mol. Diagn. 2018, 18, 165–180. [Google Scholar] [CrossRef] [PubMed]
- Bembea, M.M.; Rizkalla, N.; Freedy, J.; Barasch, N.; Vaidya, D.; Pronovost, P.J.; Everett, A.D.; Mueller, G. Plasma biomarkers of brain injury as diagnostic tools and outcome predictors after extracorporeal membrane oxygenation. Crit. Care Med. 2015, 43, 2202–2211. [Google Scholar] [CrossRef]
- Cordeiro, M.; Ferreira Carlos, F.; Pedrosa, P.; Lopez, A.; Baptista, P.V. Gold nanoparticles for diagnostics: Advances towards points of care. Diagnostics 2016, 6, 43. [Google Scholar] [CrossRef]
- Dalirirad, S.; Steckl, A.J. Lateral flow assay using aptamer-based sensing for on-site detection of dopamine in urine. Anal. Biochem. 2020, 596, 113637. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, V.; Srinivasan, S.; Singh, A.; DeRosa, M.C. An aptamer-based colorimetric lateral flow assay for the detection of human epidermal growth factor receptor 2 (HER2). Anal. Biochem. 2020, 588, 113471. [Google Scholar] [CrossRef]
- Ludovini, V.; Gori, S.; Colozza, M.; Pistola, L.; Rulli, E.; Floriani, I.; Pacifico, E.; Tofanetti, F.; Sidoni, A.; Basurto, C. Evaluation of serum HER2 extracellular domain in early breast cancer patients: Correlation with clinicopathological parameters and survival. Ann. Oncol. 2008, 19, 883–890. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Kumar, A.; Sachan, M.; Gupta, S.; Nara, S. Aptamer-gold nanozyme based competitive lateral flow assay for rapid detection of CA125 in human serum. Biosens. Bioelectron. 2020, 165, 112368. [Google Scholar] [CrossRef]
- Gao, Y.; Zhu, Z.; Xi, X.; Cao, T.; Wen, W.; Zhang, X.; Wang, S. An aptamer-based hook-effect-recognizable three-line lateral flow biosensor for rapid detection of thrombin. Biosens. Bioelectron. 2019, 133, 177–182. [Google Scholar] [CrossRef]
- Mukama, O.; Wu, W.; Wu, J.; Lu, X.; Liu, Y.; Liu, Y.; Liu, J.; Zeng, L. A highly sensitive and specific lateral flow aptasensor for the detection of human osteopontin. Talanta 2020, 210, 120624. [Google Scholar] [CrossRef] [PubMed]
- Raston, N.H.A.; Nguyen, V.-T.; Gu, M.B. A new lateral flow strip assay (LFSA) using a pair of aptamers for the detection of Vaspin. Biosens. Bioelectron. 2017, 93, 21–25. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.; Ruppert, C.; Rentschler, S.; Laufer, S.; Deigner, H.-P. Combining aptamers and antibodies: Lateral flow quantification for thrombin and interleukin-6 with smartphone readout. Sens. Actuators B Chem. 2021, 333, 129246. [Google Scholar] [CrossRef]
- Qin, C.; Gao, Y.; Wen, W.; Zhang, X.; Wang, S. Visual multiple recognition of protein biomarkers based on an array of aptamer modified gold nanoparticles in biocomputing to strip biosensor logic operations. Biosens. Bioelectron. 2016, 79, 522–530. [Google Scholar] [CrossRef]
- Xing, G.; Zhang, W.; Li, N.; Pu, Q.; Lin, J.-M. Recent progress on microfluidic biosensors for rapid detection of pathogenic bacteria. Chin. Chem. Lett. 2021. [Google Scholar] [CrossRef]
- Song, M.; Yang, M.; Hao, J. Pathogenic virus detection by optical nanobiosensors. Cell Rep. Phys. Sci. 2021, 2, 100288. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Wang, Q.; Zhang, Y. Recent advances in microchip-based methods for the detection of pathogenic bacteria. Chin. Chem. Lett. 2021. [Google Scholar] [CrossRef]
- Ren, Y.; Gao, P.; Song, Y.; Yang, X.; Yang, T.; Chen, S.; Fu, S.; Qin, X.; Shao, M.; Man, C. An aptamer-exonuclease III (Exo III)–assisted amplification-based lateral flow assay for sensitive detection of Escherichia coli O157: H7 in milk. J. Dairy Sci. 2021, 104, 8517–8529. [Google Scholar] [CrossRef]
- Tasbasi, B.B.; Guner, B.C.; Sudagidan, M.; Ucak, S.; Kavruk, M.; Ozalp, V.C. Label-free lateral flow assay for Listeria monocytogenes by aptamer-gated release of signal molecules. Anal. Biochem. 2019, 587, 113449. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lee, J.; Lee, B.H.; Song, C.-S.; Gu, M.B. Specific detection of avian influenza H5N2 whole virus particles on lateral flow strips using a pair of sandwich-type aptamers. Biosens. Bioelectron. 2019, 134, 123–129. [Google Scholar] [CrossRef]
- Le, T.T.; Chang, P.; Benton, D.J.; McCauley, J.W.; Iqbal, M.; Cass, A.E. Dual recognition element lateral flow assay toward multiplex strain specific influenza virus detection. Anal. Chem. 2017, 89, 6781–6786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Wu, Y.; Ding, L.; Huang, X.; Xiong, Y. Point-of-care COVID-19 diagnostics powered by lateral flow assay. TrAC Trends Anal. Chem. 2021, 145, 116452. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Wang, L.; He, Y.; Wang, Y.; Yang, X.; Fu, S.; Qin, X.; Chen, Q.; Man, C.; Jiang, Y. An enhanced lateral flow assay based on aptamer–magnetic separation and multifold AuNPs for ultrasensitive detection of Salmonella typhimurium in milk. Foods 2021, 10, 1605. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Gao, X.; Chen, Y.; Ren, J.; Liu, C. Aptamer-based lateral flow test strip for the simultaneous detection of Salmonella typhimurium, Escherichia coli O157:H7 and Staphylococcus aureus. Anal. Lett. 2020, 53, 646–659. [Google Scholar] [CrossRef]
- Dalirirad, S.; Steckl, A.J. Aptamer-based lateral flow assay for point of care cortisol detection in sweat. Sens. Actuators B Chem. 2019, 283, 79–86. [Google Scholar] [CrossRef]
- Dalirirad, S.; Han, D.; Steckl, A.J. Aptamer-based lateral flow biosensor for rapid detection of salivary cortisol. ACS Omega 2020, 5, 32890–32898. [Google Scholar] [CrossRef]
- Alnajrani, M.N.; Alsager, O.A. Lateral flow aptasensor for progesterone: Competitive target recognition and displacement of short complementary sequences. Anal. Biochem. 2019, 587, 113461. [Google Scholar] [CrossRef] [PubMed]
- Majdinasab, M.; Mishra, R.K.; Tang, X.; Marty, J.L. Detection of antibiotics in food: New achievements in the development of biosensors. TrAC Trends Anal. Chem. 2020, 127, 115883. [Google Scholar] [CrossRef]
- Munteanu, F.-D.; Titoiu, A.M.; Marty, J.-L.; Vasilescu, A. Detection of antibiotics and evaluation of antibacterial activity with screen-printed electrodes. Sensors 2018, 18, 901. [Google Scholar] [CrossRef] [Green Version]
- Parthasarathy, R.; Monette, C.E.; Bracero, S.; Saha, M.S. Methods for field measurement of antibiotic concentrations: Limitations and outlook. FEMS Microbiol. Ecol. 2018, 94, fiy105. [Google Scholar] [CrossRef]
- Ou, Y.; Jin, X.; Liu, J.; Tian, Y.; Zhou, N. Visual detection of kanamycin with DNA-functionalized gold nanoparticles probe in aptamer-based strip biosensor. Anal. Biochem. 2019, 587, 113432. [Google Scholar] [CrossRef]
- Birader, K.; Kumar, P.; Tammineni, Y.; Barla, J.A.; Reddy, S.; Suman, P. Colorimetric aptasensor for on-site detection of oxytetracycline antibiotic in milk. Food Chem. 2021, 356, 129659. [Google Scholar] [CrossRef]
- Kaiser, L.; Weisser, J.; Kohl, M.; Deigner, H.-P. Small molecule detection with aptamer based lateral flow assays: Applying aptamer-C-reactive protein cross-recognition for ampicillin detection. Sci. Rep. 2018, 8, 5628. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Shi, A.; Zheng, Z.; Huang, L.; Wang, Y.; Lin, H.; Lin, X. Simultaneous quantification of ampicillin and kanamycin in water samples based on lateral flow aptasensor strip with an internal line. Molecules 2021, 26, 3806. [Google Scholar] [CrossRef]
- Hassani, S.; Akmal, M.R.; Salek-Maghsoudi, A.; Rahmani, S.; Ganjali, M.R.; Norouzi, P.; Abdollahi, M. Novel label-free electrochemical aptasensor for determination of Diazinon using gold nanoparticles-modified screen-printed gold electrode. Biosens. Bioelectron. 2018, 120, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Hassani, S.; Maghsoudi, A.S.; Akmal, M.R.; Rahmani, S.R.; Sarihi, P.; Ganjali, M.R.; Norouzi, P.; Abdollahi, M. A sensitive aptamer-based biosensor for electrochemical quantification of PSA as a specific diagnostic marker of prostate cancer. J. Pharm. Pharm. Sci. 2020, 23, 243–258. [Google Scholar] [CrossRef]
- Hassani, S.; Rezaei Akmal, M.; Salek Maghsoudi, A.; Rahmani, S.; Vakhshiteh, F.; Norouzi, P.; Ganjali, M.R.; Abdollahi, M. High-performance voltammetric aptasensing platform for ultrasensitive detection of bisphenol A as an environmental pollutant. Front. Bioeng. Biotechnol. 2020, 8, 1055. [Google Scholar] [CrossRef] [PubMed]
- Maghsoudi, A.S.; Hassani, S.; Akmal, M.R.; Ganjali, M.R.; Mirnia, K.; Norouzi, P.; Abdollahi, M. An electrochemical aptasensor platform based on flower-like gold microstructure-modified screen-printed carbon electrode for detection of serpin A12 as a Type 2 diabetes biomarker. Int. J. Nanomed. 2020, 15, 2219. [Google Scholar] [CrossRef] [Green Version]





| Strategy | Biomarker | Label | Test Line | Control Line | LOD | Detection Range | Matrix | Ref. |
|---|---|---|---|---|---|---|---|---|
| Conjugation of AuNPs with dopamine duplex aptamer and dissociation of duplex in the presence of target | Dopamine | AuNPs | Streptavidin-biotinylated cDNA3 | Streptavidin-biotinylated cDNA1 | 50 ng mL−1 | nr * | Urine | [37] |
| Desorption of biotin-modified aptamer from AuNPs surface in the presence of analyte | HER2 | AuNPs | Streptavidin and pullulan mixture | Cationic charged PDDA polymer | 20 nM | nr | Human serum | [38] |
| Competitive reaction between CA125 conjugated with AuNPs and unlabeled CA125 for binding to capture probe | CA125 | AuNPs nanozyme | CA125 specific aptamer | - | 5.21 U mL−1 | 7.5–200 U mL−1 | Human serum | [40] |
| An aptamer-based hook-effect-recognizable three-line LFA | Thrombin | AuNPs | A biotinylated aptamer with poly A tail (T-DNA) | A biotinylated poly T oligonucleotide probe (C-DNA) | 0.85 nM | 1 nM–100 µM | Human serum | [41] |
| Binding of a biotinylated aptamer to target in the sample and reaction with AuNPs-streptavidin conjugate, then subsequent capturing by the antibody at the test line | Osteopontin | AuNPs | Osteopontin antibody | Complementary ssDNA | 0.1 ng mL−1 | 10–500 ng mL−1 | Human serum | [42] |
| A sandwich LFA based on biotin-labeled primary aptamer immobilized on streptavidin coated membrane as a capturing probe and secondary aptamer conjugated with AuNPs as a signaling probe. | Vaspin | AuNPs | Streptavidin-biotinylated aptamer | Streptavidin-biotinylated complementary aptamer | 0.105 nM | 0.105–25 nM | Human serum | [43] |
| Aptamer-quantum dot conjugate towards thrombin, and antibody-quantum dot conjugate against interleukin-6 | Thrombin Interleukin-6 | Green and red quantum dots | Streptavidin-biotinylated aptamer against thrombin Streptavidin- interleukin antibody | Anti-mouse antibody | 3 nM 100 pM | nr | Human serum | [44] |
| Visual multiple recognition of protein biomarkers based on an array of aptamer- modified AuNPs | Thrombin Mucin Carcinoembryonic antigen | AuNPs | T1: Streptavidin T2: streptavidin-biotinylated mucin 1 protein test DNA | Streptavidin-biotinylated thrombin control DNA | 1.61 nM 1.13 nM 0.7 nM | 3.2–250 nM 1.6–400 nM 0.8–300 nM | Human serum | [45] |
| Strategy | Microbial Analyte | Label | Test Line | Control Line | LOD | Detection Range | Matrix | Ref. |
|---|---|---|---|---|---|---|---|---|
| Combining aptamer-exonuclease III-assisted amplification with LFA | E. coli O157:H7 | AuNPs | Biotinylated poly T oligonucleotide probe | Biotinylated complementary sequence to the AuNPs probes | 7.6 × 101 cfu mL−1 8.35 × 102 cfu mL−1 | 102–106 cfu mL−1 | Culture media Milk | [49] |
| Aptamer-gated release of TMB as signal molecule and HRP activity to generate signal | L. monocytogenes | Silica nanoparticles | Immobilized HRP | No control line | 53 cells mL−1 | nr | Chicken | [50] |
| Sandwich LFA for the detection of whole virus particles | Avian influenza H5N2 | AuNPs | Biotin modified primary aptamer | Biotin modified poly A sequence | 1.2 × 106 EID50 mL−1 | 1.2 × 106 EID50 mL−1–1 × 107 EID50 mL−1 | Duck’s feces | [51] |
| A dual recognition element LFA using both aptamer and antibody | Influenza virus (strain A/H3N2/Panama/2007/99) | AuNPs | Streptavidin | Antibody | 2 × 106 virus particle | - | - | [52] |
| Pre-enrichment with magnetic nanoparticles conjugated with aptamer–ssDNA1and detection of released ssDNA1using LFA strip and AuNPs capture probe | Salmonella Typhimurium | AuNPs | Streptavidin-biotin modified ssDNA complementary with ssDNA1 | Streptavidin-biotin modified poly A ssDNA complementary with poly T ssDNA on AuNPs surface | 4.1 × 102 cfu mL−1 | 8.6 × 102–8.6 × 107 cfu mL−1 | Milk | [54] |
| Complex formation between AuNPs-aptamer1 conjugate, analyte and apatmer 2 immobilized on the test line | Salmonella Typhimurium E. coli O157:H7 Staphylococcus aureus | AuNPs | Streptavidin-biotin modified aptamer2 | Streptavidin-biotin modified complementary DNA with AuNPs-aptamer conjugate | 5 × 103 cfu mL−1 3 × 104 cfu mL−1 2 × 104 cfu mL−1 | nr | Food samples | [55] |
| Strategy | Hormone | Label | Test Line | Control Line | LOD | Detection Range | Matrix | Ref. |
|---|---|---|---|---|---|---|---|---|
| Desorption of specific aptamer from AuNPs surface in the presence of target molecule | Cortisol | AuNPs | Cysteamine | - | 1 ng mL−1 | nr | Sweat | [56] |
| Dissociation of specific aptamer from duplex probe conjugated with AuNPs in the presence of target | Cortisol | AuNPs | Biotin-modified oligonucleotide probe | Biotin-modified poly A probe | 0.37 ng mL−1 | 0.5–15 ng mL−1 | Salivary | [57] |
| Dissociation of specific aptamer from duplex probe conjugated with AuNPs in the presence of target | Progesterone | AuNPs | Streptavidin | - | 5 nM | nr | Water | [58] |
| Strategy | Antibiotic | Label | Test Line | Control Line | LOD | Detection Range | Matrix | Ref. |
|---|---|---|---|---|---|---|---|---|
| Magnetic sepration of analyte by aptamer-cDNA duplex conjugated with magnetic microspheres; then detection of cDNA by LFA strip | Kanamycin | AuNPs | Streptavidin-biotin-capture DNA1 | Streptavidin-biotin-capture DNA2 | 4.96 nm | 5–500 nM | Milk, milk products, honey | [62] |
| Competitive reaction between analyte in sample and analyte-career protein conjugate on the test line for binding to AuNPs-aptamer | Oxytetracycline | AuNPs | Oxytetracycline-carrier protein | Biotin-modified complementary probe | 5 ng mL−1 | nr | Milk | [63] |
| Competitive reaction between analyte in sample and C-reactive protein conjugated with biotin for binding to AuNPs-aptamer-mFc | Ampicillin | AuNPs | Streptavidin | α-mouse antibody | 185 mg L−1 | nr | Milk | [64] |
| Complex formation between analyte in sample, labeled DNA on conjugate pad and capture DNA on the test line | Ampicillin Kanamycin | Hexachloro-6-carboxyfluorescein | Biotin-modified ampicillin capture DNA Biotin-modified kanamycin capture DNA | - | 0.06 ng L−1 0.015 ng L−1 | 0.5–500 ng L−1 0.5–1000 ng L−1 | Water | [65] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majdinasab, M.; Badea, M.; Marty, J.L. Aptamer-Based Lateral Flow Assays: Current Trends in Clinical Diagnostic Rapid Tests. Pharmaceuticals 2022, 15, 90. https://doi.org/10.3390/ph15010090
Majdinasab M, Badea M, Marty JL. Aptamer-Based Lateral Flow Assays: Current Trends in Clinical Diagnostic Rapid Tests. Pharmaceuticals. 2022; 15(1):90. https://doi.org/10.3390/ph15010090
Chicago/Turabian StyleMajdinasab, Marjan, Mihaela Badea, and Jean Louis Marty. 2022. "Aptamer-Based Lateral Flow Assays: Current Trends in Clinical Diagnostic Rapid Tests" Pharmaceuticals 15, no. 1: 90. https://doi.org/10.3390/ph15010090