Post-Transcriptional Regulatory Crosstalk between MicroRNAs and Canonical TGF-β/BMP Signalling Cascades on Osteoblast Lineage: A Comprehensive Review
Abstract
:1. Introduction
2. MicroRNAs: A Small Molecule with Great Regulatory Functions
3. The Osteogenic Regulating Actions of MicroRNAs and Protein Catalysts Involved in MicroRNAs’ Biogenesis
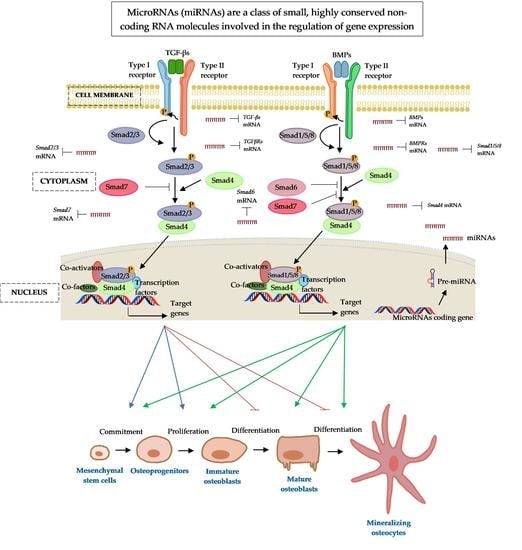
4. Transforming Growth Factor-Beta (TGF-Β)/Bone Morphogenic Protein (BMP) Signalling
5. MicroRNAs Regulating the Transforming Growth Factor-β (TGF-β) Signalling Pathway
5.1. MicroRNAs and Transforming Growth Factor-β (TGF-β) Ligands
5.2. MicroRNAs and TGF-β Receptors (TGFBR)
6. MicroRNAs Regulating the Bone Morphogenic Proteins (BMP) Signalling Pathway
6.1. MicroRNAs and Bone Morphogenic Proteins (BMP) Ligands
6.1.1. MicroRNAs and BMP-2 Ligands
6.1.2. MicroRNAs and Other Members of BMP Ligands
6.2. MicroRNAs and Bone Morphogenic Protein Receptor (BMPR)
7. MicroRNA Regulation of the Smad Cascades
7.1. MicroRNAs and the BMP-Regulated Smads (R-Smads)—Smad1 and Smad5
7.1.1. MicroRNAs and Smad1
7.1.2. MicroRNAs and Smad5
7.2. MicroRNAs and the TGF-β-Regulated Smads (R-Smads)—Smad2 and Smad3
7.2.1. MicroRNAs and Smad2
7.2.2. MicroRNAs and Smad3
7.3. MicroRNAs and the Partner or Common Smad (Co-Smad)—Smad4
7.4. MicroRNAs and the Inhibitory Smads (I-Smads)—Smad6 and Smad7
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Su, N.; Yang, J.; Xie, Y.; Du, X.; Chen, H.; Zhou, H.; Chen, L. Bone function, dysfunction and its role in diseases including critical illness. Int. J. Biol. Sci. 2019, 15, 776–787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, B. Normal bone anatomy and physiology. Clin. J. Am. Soc. Nephrol. 2008, 3 (Suppl. 3), S131–S139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berendsen, A.D.; Olsen, B.R. Bone development. Bone 2015, 80, 14–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langdahl, B.; Ferrari, S.; Dempster, D.W. Bone modeling and remodeling: Potential as therapeutic targets for the treatment of osteoporosis. Ther. Adv. Musculoskelet. Dis. 2016, 8, 225–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setiawati, S.; Rahardjo, P. Bone Development and Growth. In Osteogenesis and Bone Regeneration; Yang, H., Ed.; IntechOpen: London, UK, 2018; pp. 1–20. ISBN 978-1-83962-146-8. [Google Scholar] [CrossRef] [Green Version]
- Day, T.F.; Guo, X.; Garrett-Beal, L.; Yang, Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 2005, 8, 739–750. [Google Scholar] [CrossRef] [Green Version]
- Carroll, S.H.; Ravid, K. Differentiation of mesenchymal stem cells to osteoblasts and chondrocytes: A focus on adenosine receptors. Expert Rev. Mol. Med. 2013, 15, e1. [Google Scholar] [CrossRef] [Green Version]
- Rutkovskiy, A.; Stensløkken, K.O.; Vaage, I.J. Osteoblast differentiation at a glance. Med. Sci. Monit. Basic Res. 2016, 22, 95–106. [Google Scholar] [CrossRef] [Green Version]
- Senarath-Yapa, K.; Li, S.; Meyer, N.P.; Longaker, M.T.; Quarto, N. Integration of multiple signaling pathways determines differences in the osteogenic potential and tissue regeneration of neural crest-derived and mesoderm-derived calvarial bones. Int. J. Mol. Sci. 2013, 14, 5978–5997. [Google Scholar] [CrossRef]
- Kenkre, J.S.; Bassett, J. The bone remodelling cycle. Ann. Clin. Biochem. 2018, 55, 308–327. [Google Scholar] [CrossRef]
- Galea, G.L.; Zein, M.R.; Allen, S.; Francis-West, P. Making and shaping endochondral and intramembranous bones. Dev. Dyn. 2021, 250, 414–449. [Google Scholar] [CrossRef]
- Tani, S.; Okada, H.; Chung, U.I.; Ohba, S.; Hojo, H. The progress of stem cell technology for skeletal regeneration. Int. J. Mol. Sci. 2021, 22, 1404. [Google Scholar] [CrossRef]
- Hu, C.; Qin, Q.-H. Bone remodeling and biological effects of mechanical stimulus. AIMS Bioeng. 2020, 7, 12–28. [Google Scholar] [CrossRef]
- Katsimbri, P. The biology of normal bone remodelling. Eur. J. Cancer Care 2017, 26, e12740. [Google Scholar] [CrossRef]
- Feng, X.; McDonald, J.M. Disorders of bone remodeling. Annu. Rev. Pathol. 2011, 6, 121–145. [Google Scholar] [CrossRef] [Green Version]
- Arias, C.F.; Herrero, M.A.; Echeverri, L.F.; Oleaga, G.E.; López, J.M. Bone remodeling: A tissue-level process emerging from cell-level molecular algorithms. PLoS ONE 2018, 13, e0204171. [Google Scholar] [CrossRef] [Green Version]
- Owen, R.; Reilly, G.C. In Vitro models of bone remodelling and associated disorders. Front. Bioeng. Biotechnol. 2018, 6, 134. [Google Scholar] [CrossRef] [Green Version]
- Erben, R.G. Hypothesis: Coupling between resorption and formation in cancellous bone remodeling is a mechanically controlled event. Front. Endocrinol. 2015, 6, 82. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, J.A.; Partridge, N.C. Physiological bone remodeling: Systemic regulation and growth factor involvement. Physiology 2016, 31, 233–245. [Google Scholar] [CrossRef]
- Lerner, U.H.; Kindstedt, E.; Lundberg, P. The critical interplay between bone resorbing and bone forming cells. J. Clin. Periodontol. 2019, 46 (Suppl. 21), 33–51. [Google Scholar] [CrossRef]
- Lian, J.B.; Stein, G.S.; van Wijnen, A.J.; Stein, J.L.; Hassan, M.Q.; Gaur, T.; Zhang, Y. MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 2012, 8, 212–227. [Google Scholar] [CrossRef] [Green Version]
- Papaioannou, G.; Mirzamohammadi, F.; Kobayashi, T. MicroRNAs involved in bone formation. Cell. Mol. Life Sci. 2014, 71, 4747–4761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhaskaran, M.; Mohan, M. MicroRNAs: History, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014, 51, 759–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schickel, R.; Boyerinas, B.; Park, S.M.; Peter, M.E. MicroRNAs: Key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 2008, 27, 5959–5974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hartig, S.M.; Hamilton, M.P.; Bader, D.A.; McGuire, S.E. The miRNA interactome in metabolic homeostasis. Trends Endocrinol. Metab. 2015, 26, 733–745. [Google Scholar] [CrossRef] [Green Version]
- Mens, M.; Ghanbari, M. Cell cycle regulation of stem cells by microRNAs. Stem Cell Rev. Rep. 2018, 14, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loh, H.-Y.; Lau, Y.-Y.; Lai, K.-S.; Osman, M.A. MicroRNAs in Bone Diseases: Progress and prospects. In Transcriptional and Post-Transcriptional Regulation; Ghedira, K., Ed.; IntechOpen: London, UK, 2018; pp. 77–100. ISBN 978-1-78923-791-7. [Google Scholar] [CrossRef] [Green Version]
- Loh, H.Y.; Norman, B.P.; Lai, K.S.; Rahman, N.; Alitheen, N.; Osman, M.A. The Regulatory Role of MicroRNAs in Breast Cancer. Int. J. Mol. Sci. 2019, 20, 4940. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Moran, Y.; Agron, M.; Praher, D.; Technau, U. The evolutionary origin of plant and animal microRNAs. Nat. Ecol. Evol. 2017, 1, 27. [Google Scholar] [CrossRef] [Green Version]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358–369. [Google Scholar] [CrossRef]
- Siddika, T.; Heinemann, I.U. Bringing microRNAs to light: Methods for microRNA quantification and visualization in live cells. Front. Bioeng. Biotechnol. 2021, 8, 619583. [Google Scholar] [CrossRef]
- Chen, L.; Heikkinen, L.; Wang, C.; Yang, Y.; Sun, H.; Wong, G. Trends in the development of miRNA bioinformatics tools. Brief. Bioinform. 2019, 20, 1836–1852. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive analysis of human microRNA-mRNA interactome. Front. Genet. 2019, 10, 933. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, D. The pattern of microRNA binding site distribution. Genes 2017, 8, 296. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [Green Version]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Macfarlane, L.A.; Murphy, P.R. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [Green Version]
- Schanen, B.C.; Li, X. Transcriptional regulation of mammalian miRNA genes. Genomics 2011, 97, 1–6. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.H.; Kim, Y.K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Gregory, R.I.; Yan, K.P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Roth, B.M.; Ishimaru, D.; Hennig, M. The core microprocessor component DiGeorge syndrome critical region 8 (DGCR8) is a nonspecific RNA-binding protein. J. Biol. Chem. 2013, 288, 26785–26799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gebert, L.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Okada, C.; Yamashita, E.; Lee, S.J.; Shibata, S.; Katahira, J.; Nakagawa, A.; Yoneda, Y.; Tsukihara, T. A high-resolution structure of the pre-microRNA nuclear export machinery. Science 2009, 326, 1275–1279. [Google Scholar] [CrossRef]
- Macrae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Zhang, X.; Graves, P.; Zeng, Y. A comprehensive analysis of precursor microRNA cleavage by human Dicer. RNA 2012, 18, 2083–2092. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Doudna, J.A. TRBP alters human precursor microRNA processing In Vitro. RNA 2012, 18, 2012–2019. [Google Scholar] [CrossRef] [Green Version]
- Koscianska, E.; Starega-Roslan, J.; Krzyzosiak, W.J. The role of Dicer protein partners in the processing of microRNA precursors. PLoS ONE 2011, 6, e28548. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP complex formation ensures accurate mammalian microRNA biogenesis. Mol. Cell 2015, 57, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [Green Version]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–23. [Google Scholar] [CrossRef] [Green Version]
- Guo, L.; Lu, Z. The fate of miRNA* strand through evolutionary analysis: Implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE 2010, 5, e11387. [Google Scholar] [CrossRef] [Green Version]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. MicroRNA strand selection: Unwinding the rules. Wiley Interdiscip. Rev. RNA 2021, 12, e1627. [Google Scholar] [CrossRef]
- Felekkis, K.; Touvana, E.; Stefanou, C.H.; Deltas, C. MicroRNAs: A newly described class of encoded molecules that play a role in health and disease. Hippokratia 2010, 14, 236–240. [Google Scholar]
- Thomson, D.W.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretti, F.; Thermann, R.; Hentze, M.W. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA 2010, 16, 2493–2502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, W.; San Lucas, A.; Wang, Z.; Liu, Y. Identifying microRNA targets in different gene regions. BMC Bioinform. 2014, 15 (Suppl. 7), S4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Xue, H.; Tan, G.; Xu, Z. Effects of miRNAs, lncRNAs and circRNAs on osteoporosis as regulatory factors of bone homeostasis (Review). Mol. Med. Rep. 2021, 24, 788. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhi, H.; Zhang, Y.; Liu, Y.; Zhang, J.; Gao, Y.; Guo, M.; Ning, S.; Li, X. miRSponge: A manually curated database for experimentally supported miRNA sponges and ceRNAs. Database 2015, 2015, bav098. [Google Scholar] [CrossRef] [Green Version]
- Mirza, F.; Canalis, E. Management of endocrine disease: Secondary osteoporosis: Pathophysiology and management. Eur. J. Endocrinol. 2015, 173, R131–R151. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Guo, Q.; Wei, W.; Robertson, S.; Yuan, Y.; Luo, X. Current progress on microRNA-based gene delivery in the treatment of osteoporosis and osteoporotic fracture. Int. J. Endocrinol. 2019, 2019, 6782653. [Google Scholar] [CrossRef] [Green Version]
- Sun, Z.; Wang, H.; Wang, Y.; Yuan, G.; Yu, X.; Jiang, H.; Wu, Q.; Yang, B.; Hu, Z.; Shi, F.; et al. MiR-103-3p targets the m6A methyltransferase METTL14 to inhibit osteoblastic bone formation. Aging Cell 2021, 20, e13298. [Google Scholar] [CrossRef]
- Choi, Y.J.; Jeong, S.; Yoon, K.A.; Sung, H.J.; Cho, H.S.; Kim, D.W.; Cho, J.Y. Deficiency of DGCR8 increases bone formation through downregulation of miR-22 expression. Bone 2017, 103, 287–294. [Google Scholar] [CrossRef]
- Shirazi, S.; Huang, C.C.; Kang, M.; Lu, Y.; Ravindran, S.; Cooper, L.F. The importance of cellular and exosomal miRNAs in mesenchymal stem cell osteoblastic differentiation. Sci. Rep. 2021, 11, 5953. [Google Scholar] [CrossRef]
- Guo, L.; Xu, J.; Qi, J.; Zhang, L.; Wang, J.; Liang, J.; Qian, N.; Zhou, H.; Wei, L.; Deng, L. MicroRNA-17-92a upregulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. J. Cell Sci. 2013, 126, 978–988. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.L.; Hankenson, K.D. Integration of BMP, Wnt, and notch signaling pathways in osteoblast differentiation. J. Cell. Biochem. 2011, 112, 3491–3501. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Li, Z.; Hou, Y.; Fang, W. Potential mechanisms underlying the Runx2 induced osteogenesis of bone marrow mesenchymal stem cells. Am. J. Transl. Res. 2015, 7, 2527–2535. [Google Scholar]
- Liu, Q.; Li, M.; Wang, S.; Xiao, Z.; Xiong, Y.; Wang, G. Recent advances of osterix transcription factor in osteoblast differentiation and bone formation. Front. Cell Dev. Biol. 2020, 8, 601224. [Google Scholar] [CrossRef]
- Feng, X.H.; Derynck, R. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 2005, 21, 659–693. [Google Scholar] [CrossRef] [Green Version]
- Tamayo, E.; Alvarez, P.; Merino, R. TGFβ superfamily members as regulators of B cell development and function-implications for autoimmunity. Int. J. Mol. Sci. 2018, 19, 3928. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, M.; Derynck, R.; Miyazono, K. TGF-β and the TGF-β family: Context-dependent roles in cell and tissue physiology. Cold Spring Harb. Perspect. Biol. 2016, 8, a021873. [Google Scholar] [CrossRef] [Green Version]
- Hata, A.; Chen, Y.G. TGF-β signaling from receptors to Smads. Cold Spring Harb. Perspect. Biol. 2016, 8, a022061. [Google Scholar] [CrossRef] [Green Version]
- Massagué, J.; Seoane, J.; Wotton, D. Smad transcription factors. Genes Dev. 2005, 19, 2783–2810. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.W.; Hu, M.; Chai, J.; Seoane, J.; Huse, M.; Li, C.; Rigotti, D.J.; Kyin, S.; Muir, T.W.; Fairman, R.; et al. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-beta signaling. Mol. Cell 2001, 8, 1277–1289. [Google Scholar] [CrossRef]
- Macias, M.J.; Martin-Malpartida, P.; Massagué, J. Structural determinants of Smad function in TGF-β signaling. Trends Biochem. Sci. 2015, 40, 296–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nickel, J.; Mueller, T.D. Specification of BMP signaling. Cells 2019, 8, 1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzavlaki, K.; Moustakas, A. TGF-β Signaling. Biomolecules 2020, 10, 487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Duffhues, G.; Williams, E.; Goumans, M.J.; Heldin, C.H.; Ten Dijke, P. Bone morphogenetic protein receptors: Structure, function and targeting by selective small molecule kinase inhibitors. Bone 2020, 138, 115472. [Google Scholar] [CrossRef]
- Zhang, Y.E. Non-Smad signaling pathways of the TGF-β family. Cold Spring Harb. Perspect. Biol. 2017, 9, a022129. [Google Scholar] [CrossRef] [Green Version]
- Gazzerro, E.; Canalis, E. Bone morphogenetic proteins and their antagonists. Rev. Endocr. Metab. Disord. 2006, 7, 51–65. [Google Scholar] [CrossRef]
- Lissenberg-Thunnissen, S.N.; de Gorter, D.J.; Sier, C.F.; Schipper, I.B. Use and efficacy of bone morphogenetic proteins in fracture healing. Int. Orthop. 2011, 35, 1271–1280. [Google Scholar] [CrossRef] [Green Version]
- Ali, I.H.; Brazil, D.P. Bone morphogenetic proteins and their antagonists: Current and emerging clinical uses. Br. J. Pharmacol. 2014, 171, 3620–3632. [Google Scholar] [CrossRef] [Green Version]
- Robertson, I.B.; Rifkin, D.B. Regulation of the bioavailability of TGF-β and TGF-β-related proteins. Cold Spring Harb. Perspect. Biol. 2016, 8, a021907. [Google Scholar] [CrossRef]
- Tang, S.Y.; Alliston, T. Regulation of postnatal bone homeostasis by TGFβ. BoneKEy Rep. 2013, 2, 255. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Chen, G.; Li, Y.P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef] [Green Version]
- Grafe, I.; Alexander, S.; Peterson, J.R.; Snider, T.N.; Levi, B.; Lee, B.; Mishina, Y. TGF-β family signaling in mesenchymal differentiation. Cold Spring Harb. Perspect. Biol. 2018, 10, a022202. [Google Scholar] [CrossRef]
- Okamura, T.; Morita, K.; Iwasaki, Y.; Inoue, M.; Komai, T.; Fujio, K.; Yamamoto, K. Role of TGF-β3 in the regulation of immune responses. Clin. Exp. Rheumatol. 2015, 33 (Suppl. 92), S63–S69. [Google Scholar]
- Geiser, A.G.; Zeng, Q.Q.; Sato, M.; Helvering, L.M.; Hirano, T.; Turner, C.H. Decreased bone mass and bone elasticity in mice lacking the transforming growth factor-beta1 gene. Bone 1998, 23, 87–93. [Google Scholar] [CrossRef]
- Kinoshita, A.; Saito, T.; Tomita, H.; Makita, Y.; Yoshida, K.; Ghadami, M.; Yamada, K.; Kondo, S.; Ikegawa, S.; Nishimura, G.; et al. Domain-specific mutations in TGFB1 result in Camurati-Engelmann disease. Nat. Genet. 2000, 26, 19–20. [Google Scholar] [CrossRef]
- Dünker, N.; Krieglstein, K. Tgfbeta2-/-Tgfbeta3-/-double knockout mice display severe midline fusion defects and early embryonic lethality. Anat. Embryol. 2002, 206, 73–83. [Google Scholar] [CrossRef]
- Takeuchi, Y.; Nakayama, K.; Matsumoto, T. Differentiation and cell surface expression of transforming growth factor-beta receptors are regulated by interaction with matrix collagen in murine osteoblastic cells. J. Biol. Chem. 1996, 271, 3938–3944. [Google Scholar] [CrossRef]
- Wang, W.; Rigueur, D.; Lyons, K.M. TGFβ signaling in cartilage development and maintenance. Birth Defects Res. Part C Embryo Today Rev. 2014, 102, 37–51. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Zheng, L.; Yuan, Q.; Zhen, G.; Crane, J.L.; Zhou, X.; Cao, X. Transforming growth factor-β in stem cells and tissue homeostasis. Bone Res. 2018, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, G.; Henle, P.; Küsswetter, M.; Moghaddam, A.; Wentzensen, A.; Richter, W.; Weiss, S. TGF-beta1 as a marker of delayed fracture healing. Bone 2005, 36, 779–785. [Google Scholar] [CrossRef]
- Zhao, P.; Wang, S.; Zhou, Y.; Zheng, H.; Zhao, G. MicroRNA-185 regulates spinal cord injuries induced by thoracolumbar spine compression fractures by targeting transforming growth factor-β1. Exp. Ther. Med. 2017, 13, 1127–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, D.; Liu, J.; Shi, Q.; Mu, H.; Zhou, D. Regulatory role of microRNA-185 in the recovery process after ankle fracture. Exp. Ther. Med. 2018, 16, 3261–3267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baig, S.A.; Baig, M.N. Osteonecrosis of the femoral head: Etiology, investigations, and management. Cureus 2018, 10, e3171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, D.; Zhang, F.; Wang, B.; Liu, B.; Li, L.; Kim, S.Y.; Goodman, S.B.; Hernigou, P.; Cui, Q.; Lineaweaver, W.C.; et al. Guidelines for clinical diagnosis and treatment of osteonecrosis of the femoral head in adults (2019 version). J. Orthop. Transl. 2020, 21, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Sun, S.; Li, W.; Yuan, L.; Wang, X. Down-regulated microRNA-141 facilitates osteoblast activity and inhibits osteoclast activity to ameliorate osteonecrosis of the femoral head via up-regulating TGF-β2. Cell Cycle 2020, 19, 772–786. [Google Scholar] [CrossRef]
- Hwang, S.; Park, S.K.; Lee, H.Y.; Kim, S.W.; Lee, J.S.; Choi, E.K.; You, D.; Kim, C.S.; Suh, N. MiR-140-5p suppresses BMP2-mediated osteogenesis in undifferentiated human mesenchymal stem cells. FEBS Lett. 2014, 588, 2957–2963. [Google Scholar] [CrossRef] [Green Version]
- Fushimi, S.; Nohno, T.; Nagatsuka, H.; Katsuyama, H. Involvement of miR-140-3p in Wnt3a and TGFβ3 signaling pathways during osteoblast differentiation in MC3T3-E1 cells. Genes Cells 2018, 23, 517–527. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Zheng, Y.; Jia, L.; Li, W. Long noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells 2015, 33, 3481–3492. [Google Scholar] [CrossRef]
- Li, Z.; Hassan, M.Q.; Jafferji, M.; Aqeilan, R.I.; Garzon, R.; Croce, C.M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Correction: Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2019, 294, 10018. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Deng, C.; Li, Y.P. TGF-β and BMP signaling in osteoblast differentiation and bone formation. Int. J. Biol. Sci. 2012, 8, 272–288. [Google Scholar] [CrossRef] [Green Version]
- Seo, H.S.; Serra, R. Deletion of Tgfbr2 in Prx1-cre expressing mesenchyme results in defects in development of the long bones and joints. Dev. Biol. 2007, 310, 304–316. [Google Scholar] [CrossRef] [Green Version]
- Matsunobu, T.; Torigoe, K.; Ishikawa, M.; de Vega, S.; Kulkarni, A.B.; Iwamoto, Y.; Yamada, Y. Critical roles of the TGF-beta type I receptor ALK5 in perichondrial formation and function, cartilage integrity, and osteoblast differentiation during growth plate development. Dev. Biol. 2009, 332, 325–338. [Google Scholar] [CrossRef] [Green Version]
- Longobardi, L.; Li, T.; Myers, T.J.; O’Rear, L.; Ozkan, H.; Li, Y.; Contaldo, C.; Spagnoli, A. TGF-β type II receptor/MCP-5 axis: At the crossroad between joint and growth plate development. Dev. Cell 2012, 23, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Maeda, S.; Hayashi, M.; Komiya, S.; Imamura, T.; Miyazono, K. Endogenous TGF-beta signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J. 2004, 23, 552–563. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Dou, Q.; Ha, X. Let-7a-5p inhibits BMSCs osteogenesis in postmenopausal osteoporosis mice. Biochem. Biophys. Res. Commun. 2019, 510, 53–58. [Google Scholar] [CrossRef]
- Shen, G.Y.; Ren, H.; Huang, J.J.; Zhang, Z.D.; Zhao, W.H.; Yu, X.; Shang, Q.; Qiu, T.; Zhang, Y.Z.; Tang, J.J.; et al. Plastrum Testudinis extracts promote BMSC proliferation and osteogenic differentiation by regulating let-7f-5p and the TNFR2/PI3K/AKT signaling pathway. Cell. Physiol. Biochem. 2018, 47, 2307–2318. [Google Scholar] [CrossRef]
- Ren, H.; Yu, X.; Shen, G.; Zhang, Z.; Shang, Q.; Zhao, W.; Huang, J.; Yu, P.; Zhan, M.; Lu, Y.; et al. MiRNA-seq analysis of human vertebrae provides insight into the mechanism underlying GIOP. Bone 2019, 120, 371–386. [Google Scholar] [CrossRef]
- Shen, G.Y.; Ren, H.; Shang, Q.; Zhao, W.H.; Zhang, Z.D.; Yu, X.; Huang, J.J.; Tang, J.J.; Yang, Z.D.; Liang, D.; et al. Let-7f-5p regulates TGFBR1 in glucocorticoid-inhibited osteoblast differentiation and ameliorates glucocorticoid-induced bone loss. Int. J. Biol. Sci. 2019, 15, 2182–2197. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, Y.; Hu, Y.; Yu, C.; Panayi, A.C.; Zhou, W.; Cao, F.; Sun, Y.; Liu, M.; Liu, G.; et al. Regulatory T cell-exosomal miR-142-3p promotes angiogenesis and osteogenesis via TGFBR1/SMAD2 inhibition to accelerate fracture repair. Chem. Eng. J. 2021, 427, 131419. [Google Scholar] [CrossRef]
- Bhushan, R.; Grünhagen, J.; Becker, J.; Robinson, P.N.; Ott, C.E.; Knaus, P. MiR-181a promotes osteoblastic differentiation through repression of TGF-β signaling molecules. Int. J. Biochem. Cell Biol. 2013, 45, 696–705. [Google Scholar] [CrossRef]
- Zhang, X.; Chang, A.; Li, Y.; Gao, Y.; Wang, H.; Ma, Z.; Li, X.; Wang, B. MiR-140-5p regulates adipocyte differentiation by targeting transforming growth factor-β signaling. Sci. Rep. 2015, 5, 18118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, M.; Zheng, Y.; Dai, Y.; Chou, J.; Bian, X.; Wang, P.; Li, C.; Shen, J. MicroRNA-223 negatively regulates the osteogenic differentiation of periodontal ligament derived cells by directly targeting growth factor receptors. J. Transl. Med. 2022, 20, 465. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Guan, X.; Guo, F.; Zhou, J.; Chang, A.; Sun, B.; Cai, Y.; Ma, Z.; Dai, C.; Li, X.; et al. MiR-30e reciprocally regulates the differentiation of adipocytes and osteoblasts by directly targeting low-density lipoprotein receptor-related protein 6. Cell Death Dis. 2013, 4, e845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lowery, J.W.; Rosen, V. Bone morphogenetic protein-based therapeutic approaches. Cold Spring Harb. Perspect. Biol. 2018, 10, a022327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luu, H.H.; Song, W.X.; Luo, X.; Manning, D.; Luo, J.; Deng, Z.L.; Sharff, K.A.; Montag, A.G.; Haydon, R.C.; He, T.C. Distinct roles of bone morphogenetic proteins in osteogenic differentiation of mesenchymal stem cells. J. Orthop. Res. 2007, 25, 665–677. [Google Scholar] [CrossRef]
- Huang, Z.; Ren, P.G.; Ma, T.; Smith, R.L.; Goodman, S.B. Modulating osteogenesis of mesenchymal stem cells by modifying growth factor availability. Cytokine 2010, 51, 305–310. [Google Scholar] [CrossRef]
- Noël, D.; Gazit, D.; Bouquet, C.; Apparailly, F.; Bony, C.; Plence, P.; Millet, V.; Turgeman, G.; Perricaudet, M.; Sany, J.; et al. Short-term BMP-2 expression is sufficient for In Vivo osteochondral differentiation of mesenchymal stem cells. Stem Cells 2004, 22, 74–85. [Google Scholar] [CrossRef]
- Chan, C.K.; Seo, E.Y.; Chen, J.Y.; Lo, D.; McArdle, A.; Sinha, R.; Tevlin, R.; Seita, J.; Vincent-Tompkins, J.; Wearda, T.; et al. Identification and specification of the mouse skeletal stem cell. Cell 2015, 160, 285–298. [Google Scholar] [CrossRef] [Green Version]
- Gu, K.; Zhang, L.; Jin, T.; Rutherford, R.B. Identification of potential modifiers of Runx2/Cbfa1 activity in C2C12 cells in response to bone morphogenetic protein-7. Cells Tissues Organs 2004, 176, 28–40. [Google Scholar] [CrossRef]
- Shen, B.; Wei, A.; Whittaker, S.; Williams, L.A.; Tao, H.; Ma, D.D.; Diwan, A.D. The role of BMP-7 in chondrogenic and osteogenic differentiation of human bone marrow multipotent mesenchymal stromal cells In Vitro. J. Cell. Biochem. 2010, 109, 406–416. [Google Scholar] [CrossRef]
- Lamplot, J.D.; Qin, J.; Nan, G.; Wang, J.; Liu, X.; Yin, L.; Tomal, J.; Li, R.; Shui, W.; Zhang, H.; et al. BMP9 signaling in stem cell differentiation and osteogenesis. Am. J. Stem Cells 2013, 2, 1–21. [Google Scholar]
- Bandyopadhyay, A.; Tsuji, K.; Cox, K.; Harfe, B.D.; Rosen, V.; Tabin, C.J. Genetic analysis of the roles of BMP2, BMP4, and BMP7 in limb patterning and skeletogenesis. PLoS Genet. 2006, 2, e216. [Google Scholar] [CrossRef] [Green Version]
- Luo, G.; Hofmann, C.; Bronckers, A.L.; Sohocki, M.; Bradley, A.; Karsenty, G. BMP-7 is an inducer of nephrogenesis, and is also required for eye development and skeletal patterning. Genes Dev. 1995, 9, 2808–2820. [Google Scholar] [CrossRef] [Green Version]
- Kokabu, S.; Gamer, L.; Cox, K.; Lowery, J.; Tsuji, K.; Raz, R.; Economides, A.; Katagiri, T.; Rosen, V. BMP3 suppresses osteoblast differentiation of bone marrow stromal cells via interaction with Acvr2b. Mol. Endocrinol. 2012, 26, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Bahamonde, M.E.; Lyons, K.M. BMP3: To be or not to be a BMP. J. Bone Jt. Surg. 2001, 83 (Suppl. 1), S56–S62. [Google Scholar] [CrossRef]
- Gautschi, O.P.; Frey, S.P.; Zellweger, R. Bone morphogenetic proteins in clinical applications. ANZ J. Surg. 2007, 77, 626–631. [Google Scholar] [CrossRef]
- Grünhagen, J.; Bhushan, R.; Degenkolbe, E.; Jäger, M.; Knaus, P.; Mundlos, S.; Robinson, P.N.; Ott, C.E. MiR-497~195 cluster microRNAs regulate osteoblast differentiation by targeting BMP signaling. J. Bone Miner. Res. 2015, 30, 796–808. [Google Scholar] [CrossRef]
- Zhang, J.F.; Fu, W.M.; He, M.L.; Xie, W.D.; Lv, Q.; Wan, G.; Li, G.; Wang, H.; Lu, G.; Hu, X.; et al. MiRNA-20a promotes osteogenic differentiation of human mesenchymal stem cells by co-regulating BMP signaling. RNA Biol. 2011, 8, 829–838. [Google Scholar] [CrossRef] [Green Version]
- Akune, T.; Ohba, S.; Kamekura, S.; Yamaguchi, M.; Chung, U.I.; Kubota, N.; Terauchi, Y.; Harada, Y.; Azuma, Y.; Nakamura, K.; et al. PPARgamma insufficiency enhances osteogenesis through osteoblast formation from bone marrow progenitors. J. Clin. Investig. 2004, 113, 846–855. [Google Scholar] [CrossRef] [Green Version]
- Takano, M.; Otsuka, F.; Matsumoto, Y.; Inagaki, K.; Takeda, M.; Nakamura, E.; Tsukamoto, N.; Miyoshi, T.; Sada, K.E.; Makino, H. Peroxisome proliferator-activated receptor activity is involved in the osteoblastic differentiation regulated by bone morphogenetic proteins and tumor necrosis factor-α. Mol. Cell. Endocrinol. 2012, 348, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Cen, X.; Pan, X.; Zhang, B.; Huang, W.; Pei, F.; Luo, T.; Huang, X.; Liu, J.; Zhao, Z. MiR-20a-5p contributes to osteogenic differentiation of human dental pulp stem cells by regulating BAMBI and activating the phosphorylation of Smad5 and p38. Stem Cell Res. Ther. 2021, 12, 421. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zhang, Z.; Peng, T.; Wang, G.; Xu, Q.; Li, G. MiR-204 inhibits the osteogenic differentiation of mesenchymal stem cells by targeting bone morphogenetic protein 2. Mol. Med. Rep. 2020, 21, 43–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, G.P.; Zhang, J.; Zhu, C.H.; Lin, L.; Wang, J.; Zhang, H.J.; Li, J.; Yu, X.G.; Zhao, Z.S.; Dong, W.; et al. MicroRNA-98 regulates osteogenic differentiation of human bone mesenchymal stromal cells by targeting BMP2. J. Cell. Mol. Med. 2017, 21, 254–264. [Google Scholar] [CrossRef]
- Zheng, F.; Zhang, F.; Wang, F. Inhibition of miR-98-5p promotes high glucose-induced suppression of preosteoblast proliferation and differentiation via the activation of the PI3K/AKT/GSK3β signaling pathway by targeting BMP2. Mol. Med. Rep. 2022, 26, 292. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.G.; Shi, P.; Sun, Y.J.; Liu, H.Z.; Ni, J.Q.; Wang, X. MiR-214-3p delays fracture healing in rats with osteoporotic fracture through inhibiting BMP/Smad signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 449–455. [Google Scholar] [CrossRef]
- Zhang, H.; Ding, W.; Ji, F.; Wu, D. MicroRNA-410 participates in the pathological process of postmenopausal osteoporosis by downregulating bone morphogenetic protein-2. Exp. Ther. Med. 2019, 18, 3659–3666. [Google Scholar] [CrossRef]
- Lukasiewicz, A.M.; Bohl, D.D.; Varthi, A.G.; Basques, B.A.; Webb, M.L.; Samuel, A.M.; Grauer, J.N. Spinal fracture in patients with ankylosing spondylitis: Cohort definition, distribution of injuries, and hospital outcomes. Spine 2016, 41, 191–196. [Google Scholar] [CrossRef]
- Ding, L.; Yin, Y.; Hou, Y.; Jiang, H.; Zhang, J.; Dai, Z.; Zhang, G. MicroRNA-214-3p suppresses ankylosing spondylitis fibroblast osteogenesis via BMP-TGFβ Axis and BMP2. Front. Endocrinol. 2021, 11, 609753. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, Q.S.; Ding, W.B.; Zhang, L.L.; Wang, H.C.; Zhu, Y.J.; He, W.; Chai, Y.N.; Liu, Y.W. Increased microRNA-93-5p inhibits osteogenic differentiation by targeting bone morphogenetic protein-2. PLoS ONE 2017, 12, e0182678. [Google Scholar] [CrossRef] [Green Version]
- Ross, J.R.; Gardner, M.J. Femoral head fractures. Curr. Rev. Musculoskelet. Med. 2012, 5, 199–205. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.; Yang, X.; Tan, S.; Song, L. Author correction: Enhancing cell proliferation and osteogenic differentiation of MC3T3-E1 pre-osteoblasts by BMP-2 delivery in graphene oxide-incorporated PLGA/HA biodegradable microcarriers. Sci. Rep. 2020, 10, 6249. [Google Scholar] [CrossRef] [Green Version]
- Fu, L.; Min, N.; Cai, X. MicroRNA-543 inhibits osteogenic proliferation and differentiation by targeting bone morphogenetic protein-2. J. Biomater. Tissue Eng. 2019, 9, 198–205. [Google Scholar] [CrossRef]
- Luo, B.; Yang, J.; Yuan, Y.; Hao, P.; Cheng, X. MicroRNA-142 regulates osteoblast differentiation and apoptosis of mouse pre-osteoblast cells by targeting bone morphogenetic protein 2. FEBS Open Bio 2020, 10, 1793–1801. [Google Scholar] [CrossRef]
- Li, X.; Li, K.; Yu, G.; Yu, C.; Liu, C. MiR-342-5p inhibits expression of Bmp7 to regulate proliferation, differentiation and migration of osteoblasts. Mol. Immunol. 2019, 114, 251–259. [Google Scholar] [CrossRef]
- Wang, C.G.; Liao, Z.; Xiao, H.; Liu, H.; Hu, Y.H.; Liao, Q.D.; Zhong, D. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp. Mol. Pathol. 2019, 107, 77–84. [Google Scholar] [CrossRef]
- Zhang, N.; Hu, X.; He, S.; Ding, W.; Wang, F.; Zhao, Y.; Huang, Z. LncRNA MSC-AS1 promotes osteogenic differentiation and alleviates osteoporosis through sponging microRNA-140-5p to upregulate BMP2. Biochem. Biophys. Res. Commun. 2019, 519, 790–796. [Google Scholar] [CrossRef]
- Xiong, Y.; Chen, L.; Yan, C.; Endo, Y.; Mi, B.; Liu, G. The lncRNA Rhno1/miR-6979-5p/BMP2 axis modulates osteoblast differentiation. Int. J. Biol. Sci. 2020, 16, 1604–1615. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.D.; Han, W.X.; Liu, Y.X. Suppression of miR-451a accelerates osteogenic differentiation and inhibits bone loss via Bmp6 signaling during osteoporosis. Biomed. Pharmacother. 2019, 120, 109378. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, C.; Wu, C.; Liu, J.; Yu, H.; Zhou, X.; Zhang, J.; Wang, X.; He, S.; Xu, X.; et al. MiR-765 inhibits the osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting BMP6 via regulating the BMP6/Smad1/5/9 signaling pathway. Stem Cell Res. Ther. 2020, 11, 62. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Fu, D. LncRNA ZNF710-AS1 acts as a ceRNA for miR-146a-5p and miR-146b-5p to accelerate osteogenic differentiation of PDLSCs by upregulating the BMP6/Smad1/5/9 pathway. J. Hard Tissue Biol. 2022, 31, 231–244. [Google Scholar] [CrossRef]
- Kureel, J.; Dixit, M.; Tyagi, A.M.; Mansoori, M.N.; Srivastava, K.; Raghuvanshi, A.; Maurya, R.; Trivedi, R.; Goel, A.; Singh, D. MiR-542-3p suppresses osteoblast cell proliferation and differentiation, targets BMP-7 signaling and inhibits bone formation. Cell Death Dis. 2014, 5, e1050. [Google Scholar] [CrossRef] [PubMed]
- Asila, A.; Yang, X.; Kaisaer, Y.; Ma, L. SNHG16/miR-485-5p/BMP7 axis modulates osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. J. Gene Med. 2021, 23, e3296. [Google Scholar] [CrossRef] [PubMed]
- Tao, G.; Mao, P.; Guan, H.; Jiang, M.; Chu, T.; Zhong, C.; Liu, J. Effect of miR-181a-3p on osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by targeting BMP10. Artif. Cells Nanomed. Biotechnol. 2019, 47, 4159–4164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, L.; Fan, J.; Liu, Y.; Li, T.; Xu, H.; Yang, Y.; Deng, L.; Li, H.; Zhao, R.C. MiR-450b promotes osteogenic differentiation In Vitro and enhances bone formation In Vivo by targeting BMP3. Stem Cells Dev. 2018, 27, 600–611. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Hu, J.; Ren, W.; Fang, Y.; Hu, K.; Yu, H.; Liao, D.; Liu, S.; Zhou, L.; He, T.; et al. LncRNA SNHG3 regulates the BMSC osteogenic differentiation in bone metastasis of breast cancer by modulating the miR-1273g-3p/BMP3 axis. Biochem. Biophys. Res. Commun. 2022, 594, 117–123. [Google Scholar] [CrossRef]
- Suva, L.J.; Washam, C.; Nicholas, R.W.; Griffin, R.J. Bone metastasis: Mechanisms and therapeutic opportunities. Nat. Rev. Endocrinol. 2011, 7, 208–218. [Google Scholar] [CrossRef]
- Macedo, F.; Ladeira, K.; Pinho, F.; Saraiva, N.; Bonito, N.; Pinto, L.; Goncalves, F. Bone metastases: An overview. Oncol. Rev. 2017, 11, 321. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.S.; Akhtar, N.; Jamil, H.M.; Banik, R.S.; Asaduzzaman, S.M. TGF-β/BMP signaling and other molecular events: Regulation of osteoblastogenesis and bone formation. Bone Res. 2015, 3, 15005. [Google Scholar] [CrossRef] [Green Version]
- Mishina, Y.; Starbuck, M.W.; Gentile, M.A.; Fukuda, T.; Kasparcova, V.; Seedor, J.G.; Hanks, M.C.; Amling, M.; Pinero, G.J.; Harada, S.; et al. Bone morphogenetic protein type IA receptor signaling regulates postnatal osteoblast function and bone remodeling. J. Biol. Chem. 2004, 279, 27560–27566. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Yang, L.; Wan, M.; Cao, X. Generation of a mouse model with expression of bone morphogenetic protein type II receptor lacking the cytoplasmic domain in osteoblasts. Ann. N. Y. Acad. Sci. 2010, 1192, 286–291. [Google Scholar] [CrossRef]
- Zeng, Y.; Qu, X.; Li, H.; Huang, S.; Wang, S.; Xu, Q.; Lin, R.; Han, Q.; Li, J.; Zhao, R.C. MicroRNA-100 regulates osteogenic differentiation of human adipose-derived mesenchymal stem cells by targeting BMPR2. FEBS Lett. 2012, 586, 2375–2381. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; LV, Q.; LV, C. MicroRNA-153 suppresses the osteogenic differentiation of human mesenchymal stem cells by targeting bone morphogenetic protein receptor type II. Int. J. Mol. Med. 2015, 36, 760–766. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Zhong, L.; Yuan, T.; Chen, S.; Zhou, Y.; An, L.; Guo, Y.; Fan, M.; Li, Y.; Sun, Y.; et al. MicroRNA-155 inhibits the osteogenic differentiation of mesenchymal stem cells induced by BMP9 via downregulation of BMP signaling pathway. Int. J. Mol. Med. 2018, 41, 3379–3393. [Google Scholar] [CrossRef] [Green Version]
- Qin, W.; Liu, L.; Wang, Y.; Wang, Z.; Yang, A.; Wang, T. MiR-494 inhibits osteoblast differentiation by regulating BMP signaling in simulated microgravity. Endocrine 2019, 65, 426–439. [Google Scholar] [CrossRef]
- John, A.A.; Prakash, R.; Kureel, J.; Singh, D. Identification of novel microRNA inhibiting actin cytoskeletal rearrangement thereby suppressing osteoblast differentiation. J. Mol. Med. 2018, 96, 427–444. [Google Scholar] [CrossRef]
- Chang, M.; Lin, H.; Fu, H.; Wang, B.; Han, G.; Fan, M. MicroRNA-195-5p regulates osteogenic differentiation of periodontal ligament cells under mechanical loading. J. Cell. Physiol. 2017, 232, 3762–3774. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, S.; Yuan, S.; Zhang, H.; Liu, J. MicroRNA-23a inhibits osteogenesis of periodontal mesenchymal stem cells by targeting bone morphogenetic protein signaling. Arch. Oral Biol. 2019, 102, 93–100. [Google Scholar] [CrossRef]
- Wang, H.; Xie, Z.; Hou, T.; Li, Z.; Huang, K.; Gong, J.; Zhou, W.; Tang, K.; Xu, J.; Dong, S. MiR-125b regulates the osteogenic differentiation of human mesenchymal stem cells by targeting BMPR1b. Cell. Physiol. Biochem. 2017, 41, 530–542. [Google Scholar] [CrossRef]
- Peng, J.; Zhan, Y.; Zong, Y. METTL3-mediated LINC00657 promotes osteogenic differentiation of mesenchymal stem cells via miR-144-3p/BMPR1B axis. Cell Tissue Res. 2022, 388, 301–312. [Google Scholar] [CrossRef]
- Xie, H.; Liu, M.; Jin, Y.; Lin, H.; Zhang, Y.; Zheng, S. MiR-1323 suppresses bone mesenchymal stromal cell osteogenesis and fracture healing via inhibiting BMP4/SMAD4 signaling. J. Orthop. Surg. Res. 2020, 15, 237. [Google Scholar] [CrossRef]
- Zou, M.L.; Chen, Z.H.; Teng, Y.Y.; Liu, S.Y.; Jia, Y.; Zhang, K.W.; Sun, Z.L.; Wu, J.J.; Yuan, Z.D.; Feng, Y.; et al. The Smad dependent TGF-β and BMP signaling pathway in bone remodeling and therapies. Front. Mol. Biosci. 2021, 8, 593310. [Google Scholar] [CrossRef] [PubMed]
- Afzal, F.; Pratap, J.; Ito, K.; Ito, Y.; Stein, J.L.; van Wijnen, A.J.; Stein, G.S.; Lian, J.B.; Javed, A. Smad function and intranuclear targeting share a Runx2 motif required for osteogenic lineage induction and BMP2 responsive transcription. J. Cell. Physiol. 2005, 204, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Liang, W.; Lin, M.; Li, X.; Li, C.; Gao, B.; Gan, H.; Yang, Z.; Lin, X.; Liao, L.; Yang, M. Icariin promotes bone formation via the BMP-2/Smad4 signal transduction pathway in the hFOB 1.19 human osteoblastic cell line. Int. J. Mol. Med. 2012, 30, 889–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.M.; Lee, E.H. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng. Part B Rev. 2013, 19, 254–263. [Google Scholar] [CrossRef] [Green Version]
- Amarasekara, D.S.; Kim, S.; Rho, J. Regulation of osteoblast differentiation by cytokine networks. Int. J. Mol. Sci. 2021, 22, 2851. [Google Scholar] [CrossRef]
- Wang, M.; Jin, H.; Tang, D.; Huang, S.; Zuscik, M.J.; Chen, D. Smad1 plays an essential role in bone development and postnatal bone formation. Osteoarthr. Cartil. 2011, 19, 751–762. [Google Scholar] [CrossRef] [Green Version]
- Retting, K.N.; Song, B.; Yoon, B.S.; Lyons, K.M. BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 2009, 136, 1093–1104. [Google Scholar] [CrossRef] [Green Version]
- Keller, B.; Yang, T.; Chen, Y.; Munivez, E.; Bertin, T.; Zabel, B.; Lee, B. Interaction of TGFβ and BMP signaling pathways during chondrogenesis. PLoS ONE 2011, 6, e16421. [Google Scholar] [CrossRef] [Green Version]
- Luzi, E.; Marini, F.; Sala, S.C.; Tognarini, I.; Galli, G.; Brandi, M.L. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting of the SMAD1 transcription factor. J. Bone Miner. Res. 2008, 23, 287–295. [Google Scholar] [CrossRef]
- Su, X.; Liao, L.; Shuai, Y.; Jing, H.; Liu, S.; Zhou, H.; Liu, Y.; Jin, Y. MiR-26a functions oppositely in osteogenic differentiation of BMSCs and ADSCs depending on distinct activation and roles of Wnt and BMP signaling pathway. Cell Death Dis. 2015, 6, e1851. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Hassan, M.Q.; Volinia, S.; van Wijnen, A.J.; Stein, J.L.; Croce, C.M.; Lian, J.B.; Stein, G.S. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. USA 2008, 105, 13906–13911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, T.; Zhou, H.; Hong, Y.; Li, J.; Jiang, X.; Huang, H. MiR-30 family members negatively regulate osteoblast differentiation. J. Biol. Chem. 2012, 287, 7503–7511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyan, B.D.; Weesner, T.C.; Lohmann, C.H.; Andreacchio, D.; Carnes, D.L.; Dean, D.D.; Cochran, D.L.; Schwartz, Z. Porcine fetal enamel matrix derivative enhances bone formation induced by demineralized freeze dried bone allograft In Vivo. J. Periodontol. 2000, 71, 1278–1286. [Google Scholar] [CrossRef] [PubMed]
- Goda, S.; Inoue, H.; Kaneshita, Y.; Nagano, Y.; Ikeo, T.; Iida, J.; Domae, N. Emdogain stimulates matrix degradation by osteoblasts. J. Dent. Res. 2008, 87, 782–787. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.L.; Pan, H.X.; Zhao, B.; Dong, B.C.; Shao, L.; Fu, G.S.; Wang, Q.; Li, M. MicroRNA-100 inhibits bone morphogenetic protein-induced osteoblast differentiation by targeting Smad1. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3911–3919. [Google Scholar]
- Nemtoi, A.; Ladunca, O.; Dragan, E.; Budacu, C.; Mihai, C.; Haba, D. Quantitative and qualitative bone assessment of the posterior mandible in patients with diabetes mellitus: A cone beam computed tomography study. Rev. Med. Chir. A Soc. De Med. Si Nat. Din Iasi 2013, 117, 1002–1008. [Google Scholar]
- Clementini, M.; Rossetti, P.H.; Penarrocha, D.; Micarelli, C.; Bonachela, W.C.; Canullo, L. Systemic risk factors for peri-implant bone loss: A systematic review and meta-analysis. Int. J. Oral Maxillofac. Surg. 2014, 43, 323–334. [Google Scholar] [CrossRef]
- de Morais, J.A.; Trindade-Suedam, I.K.; Pepato, M.T.; Marcantonio, E., Jr.; Wenzel, A.; Scaf, G. Effect of diabetes mellitus and insulin therapy on bone density around osseointegrated dental implants: A digital subtraction radiography study in rats. Clin. Oral Implant. Res. 2009, 20, 796–801. [Google Scholar] [CrossRef]
- Tang, Y.; Zheng, L.; Zhou, J.; Chen, Y.; Yang, L.; Deng, F.; Hu, Y. MiR-203-3p participates in the suppression of diabetes-associated osteogenesis in the jaw bone through targeting Smad1. Int. J. Mol. Med. 2018, 41, 1595–1607. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Guo, D.; Yang, S.; Sun, H.; Wu, B.; Zhou, D. Inhibition of miR-222-3p activity promoted osteogenic differentiation of hBMSCs by regulating Smad5-RUNX2 signal axis. Biochem. Biophys. Res. Commun. 2016, 470, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Luo, Y.; Wang, J.; Zhang, N.; Gu, C.; Li, L.; Qian, D.; Cai, W.; Fan, J.; Yin, G. Exosomal miRNA-128-3p from mesenchymal stem cells of aged rats regulates osteogenesis and bone fracture healing by targeting Smad5. J. Nanobiotechnol. 2020, 18, 47. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.L.; Wei, X.; Wang, A.G.; Bai, Y.; Wu, X.J. KCNQ1OT1 regulates osteogenic differentiation of hBMSC by miR-320a/Smad5 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2843–2854. [Google Scholar] [CrossRef]
- Iaculli, F.; Di Filippo, E.S.; Piattelli, A.; Mancinelli, R.; Fulle, S. Dental pulp stem cells grown on dental implant titanium surfaces: An In Vitro evaluation of differentiation and microRNAs expression. J. Biomed. Mater. Res. Part B Appl. Biomater. 2017, 105, 953–965. [Google Scholar] [CrossRef]
- Wei, F.; Yang, S.; Guo, Q.; Zhang, X.; Ren, D.; Lv, T.; Xu, X. MicroRNA-21 regulates osteogenic differentiation of periodontal ligament stem cells by targeting Smad5. Sci. Rep. 2017, 7, 16608. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Sun, Y.; Cao, S.; Zhang, J.; Wei, J. Downregulation of miR-24-3p promotes osteogenic differentiation of human periodontal ligament stem cells by targeting SMAD family member 5. J. Cell. Physiol. 2019, 234, 7411–7419. [Google Scholar] [CrossRef]
- Fang, T.; Wu, Q.; Zhou, L.; Mu, S.; Fu, Q. MiR-106b-5p and miR-17-5p suppress osteogenic differentiation by targeting Smad5 and inhibit bone formation. Exp. Cell Res. 2016, 347, 74–82. [Google Scholar] [CrossRef]
- Gu, Y.; Ma, L.; Song, L.; Li, X.; Chen, D.; Bai, X. MiR-155 inhibits mouse osteoblast differentiation by suppressing SMAD5 expression. BioMed Res. Int. 2017, 2017, 1893520. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Sun, F.; Feng, Y.; Sun, X.; Li, J.; Fan, Q.; Liu, M. MicroRNA-132-3p represses Smad5 in MC3T3-E1 osteoblastic cells under cyclic tensile stress. Mol. Cell. Biochem. 2019, 458, 143–157. [Google Scholar] [CrossRef]
- Ellur, G.; Sukhdeo, S.V.; Khan, M.T.; Sharan, K. Maternal high protein-diet programs impairment of offspring’s bone mass through miR-24-1-5p mediated targeting of SMAD5 in osteoblasts. Cell. Mol. Life Sci. 2021, 78, 1729–1744. [Google Scholar] [CrossRef]
- Li, J.; Tsuji, K.; Komori, T.; Miyazono, K.; Wrana, J.L.; Ito, Y.; Nifuji, A.; Noda, M. Smad2 overexpression enhances Smad4 gene expression and suppresses CBFA1 gene expression in osteoblastic osteosarcoma ROS17/2.8 cells and primary rat calvaria cells. J. Biol. Chem. 1998, 273, 31009–31015. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.S.; Alliston, T.; Delston, R.; Derynck, R. Repression of Runx2 function by TGF-beta through recruitment of class II histone deacetylases by Smad3. EMBO J. 2005, 24, 2543–2555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, Y.; Xie, H.; Jin, Q.; Zhao, X.; Shi, Y.; Zhou, Y.; Hu, Z.; Ye, Y.; Huang, X.; Sun, Y.; et al. Extracellular vesicles derived from neural EGFL-Like 1-modified mesenchymal stem cells improve acellular bone regeneration via the miR-25-5p-SMAD2 signaling axis. Bioact. Mater. 2022, 17, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Fan, J.; Fan, L.; Li, T.; Yang, Y.; Xu, H.; Deng, L.; Li, J.; Li, T.; Weng, X.; et al. MiRNA-10b reciprocally stimulates osteogenesis and inhibits adipogenesis partly through the TGF-β/SMAD2 signaling pathway. Aging Dis. 2018, 9, 1058–1073. [Google Scholar] [CrossRef] [Green Version]
- Qiu, J.; Huang, G.; Na, N.; Chen, L. MicroRNA-214-5p/TGF-β/Smad2 signaling alters adipogenic differentiation of bone marrow stem cells in postmenopausal osteoporosis. Mol. Med. Rep. 2018, 17, 6301–6310. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.L.; Liu, L.; Su, Y.W.; Xian, C.J. MiR-6315 attenuates methotrexate treatment-induced decreased osteogenesis and increased adipogenesis potentially through modulating TGF-β/Smad2 signalling. Biomedicines 2021, 9, 1926. [Google Scholar] [CrossRef]
- Georgiou, K.R.; Scherer, M.A.; Fan, C.M.; Cool, J.C.; King, T.J.; Foster, B.K.; Xian, C.J. Methotrexate chemotherapy reduces osteogenesis but increases adipogenic potential in the bone marrow. J. Cell. Physiol. 2012, 227, 909–918. [Google Scholar] [CrossRef]
- Yang, X.; Chen, L.; Xu, X.; Li, C.; Huang, C.; Deng, C.X. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J. Cell Biol. 2001, 153, 35–46. [Google Scholar] [CrossRef] [Green Version]
- Cheung, K.S.; Sposito, N.; Stumpf, P.S.; Wilson, D.I.; Sanchez-Elsner, T.; Oreffo, R.O. MicroRNA-146a regulates human foetal femur derived skeletal stem cell differentiation by down-regulating SMAD2 and SMAD3. PLoS ONE 2014, 9, e98063. [Google Scholar] [CrossRef] [Green Version]
- Cui, Q.; Jo, W.L.; Koo, K.H.; Cheng, E.Y.; Drescher, W.; Goodman, S.B.; Ha, Y.C.; Hernigou, P.; Jones, L.C.; Kim, S.Y.; et al. ARCO consensus on the pathogenesis of non-traumatic osteonecrosis of the femoral head. J. Korean Med. Sci. 2021, 36, e65. [Google Scholar] [CrossRef]
- Moya-Angeler, J.; Gianakos, A.L.; Villa, J.C.; Ni, A.; Lane, J.M. Current concepts on osteonecrosis of the femoral head. World J. Orthop. 2015, 6, 590–601. [Google Scholar] [CrossRef]
- Petek, D.; Hannouche, D.; Suva, D. Osteonecrosis of the femoral head: Pathophysiology and current concepts of treatment. EFORT Open Rev. 2019, 4, 85–97. [Google Scholar] [CrossRef]
- Hao, C.; Yang, S.; Xu, W.; Shen, J.K.; Ye, S.; Liu, X.; Dong, Z.; Xiao, B.; Feng, Y. MiR-708 promotes steroid-induced osteonecrosis of femoral head, suppresses osteogenic differentiation by targeting SMAD3. Sci. Rep. 2016, 6, 22599. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Hu, J.Z.; Shi, Z.Y. MiR-181d promotes steroid-induced osteonecrosis of the femoral head by targeting SMAD3 to inhibit osteogenic differentiation of hBMSCs. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4053–4062. [Google Scholar] [CrossRef]
- Fu, L.; Liu, H.; Lei, W. MiR-596 inhibits osteoblastic differentiation and cell proliferation by targeting Smad3 in steroid-induced osteonecrosis of femoral head. J. Orthop. Surg. Res. 2020, 15, 173. [Google Scholar] [CrossRef]
- Wei, B.; Wei, W. Identification of aberrantly expressed of serum microRNAs in patients with hormone-induced non-traumatic osteonecrosis of the femoral head. Biomed. Pharmacother. 2015, 75, 191–195. [Google Scholar] [CrossRef]
- Zheng, J.; Yan, X.; Zeng, Y. MiR-423-5p downregulates osteoblastic differentiation and cell viability by targeting SMAD3 in non-traumatic osteonecrosis. Trop. J. Pharm. Res. 2021, 20, 567–572. [Google Scholar] [CrossRef]
- Reikerås, O.; Shegarfi, H.; Wang, J.E.; Utvåg, S.E. Lipopolysaccharide impairs fracture healing: An experimental study in rats. Acta Orthop. 2005, 76, 749–753. [Google Scholar] [CrossRef]
- Sakuma, Y.; Tanaka, K.; Suda, M.; Komatsu, Y.; Yasoda, A.; Miura, M.; Ozasa, A.; Narumiya, S.; Sugimoto, Y.; Ichikawa, A.; et al. Impaired bone resorption by lipopolysaccharide In Vivo in mice deficient in the prostaglandin E receptor EP4 subtype. Infect. Immun. 2000, 68, 6819–6825. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Hao, W.; Wang, X.; Su, H. MiR-23b targets Smad 3 and ameliorates the LPS-inhibited osteogenic differentiation in preosteoblast MC3T3-E1 cells. J. Toxicol. Sci. 2016, 41, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Kaur, T.; John, A.A.; Sharma, C.; Vashisht, N.K.; Singh, D.; Kapila, R.; Kapila, S. MiR300 intervenes Smad3/β-catenin/RunX2 crosstalk for therapy with an alternate function as indicative biomarker in osteoporosis. Bone 2021, 143, 115603. [Google Scholar] [CrossRef]
- Salazar, V.S.; Zarkadis, N.; Huang, L.; Norris, J.; Grimston, S.K.; Mbalaviele, G.; Civitelli, R. Embryonic ablation of osteoblast Smad4 interrupts matrix synthesis in response to canonical Wnt signaling and causes an osteogenesis-imperfecta-like phenotype. J. Cell Sci. 2013, 126 Pt 21, 4974–4984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Cao, X.; Chen, J.; Gu, J.; Zhao, J.; Sun, J. MicroRNA-224 suppresses osteoblast differentiation by inhibiting SMAD4. J. Cell. Physiol. 2018, 233, 6929–6937. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jiang, C.; Wang, X.; Wang, H.; Yan, Z.; Yuan, H. Reciprocal effect of microRNA-224 on osteogenesis and adipogenesis in steroid-induced osteonecrosis of the femoral head. Bone 2021, 145, 115844. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Kim, M.; Song, N.J.; Kim, J.H.; Seo, D.; Lee, J.H.; Jung, S.M.; Lee, J.Y.; Lee, J.; Lee, Y.S.; et al. A reciprocal role of the Smad4-Taz Axis in osteogenesis and adipogenesis of mesenchymal stem cells. Stem Cells 2019, 37, 368–381. [Google Scholar] [CrossRef] [Green Version]
- Kuang, W.; Zheng, L.; Xu, X.; Lin, Y.; Lin, J.; Wu, J.; Tan, J. Dysregulation of the miR-146a-Smad4 axis impairs osteogenesis of bone mesenchymal stem cells under inflammation. Bone Res. 2017, 5, 17037. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Geng, J.; Wei, X.; Zhang, R.; Jiang, S. MiR-144-3p regulates osteogenic differentiation and proliferation of murine mesenchymal stem cells by specifically targeting Smad4. FEBS Lett. 2016, 590, 795–807. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Jin, Y.; Hong, F.; Ma, Y.; Yang, J.; Tang, Y.; Zhu, Z.; Wu, J.; Bao, Q.; Li, L.; et al. MiR-664-3p suppresses osteoblast differentiation and impairs bone formation via targeting Smad4 and Osterix. J. Cell. Mol. Med. 2021, 25, 5025–5037. [Google Scholar] [CrossRef]
- Miyazawa, K.; Miyazono, K. Regulation of TGF-β family signaling by inhibitory Smads. Cold Spring Harb. Perspect. Biol. 2017, 9, a022095. [Google Scholar] [CrossRef] [Green Version]
- Goto, K.; Kamiya, Y.; Imamura, T.; Miyazono, K.; Miyazawa, K. Selective inhibitory effects of Smad6 on bone morphogenetic protein type I receptors. J. Biol. Chem. 2007, 282, 20603–20611. [Google Scholar] [CrossRef] [Green Version]
- Hanyu, A.; Ishidou, Y.; Ebisawa, T.; Shimanuki, T.; Imamura, T.; Miyazono, K. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J. Cell Biol. 2001, 155, 1017–1027. [Google Scholar] [CrossRef] [Green Version]
- Kavsak, P.; Rasmussen, R.K.; Causing, C.G.; Bonni, S.; Zhu, H.; Thomsen, G.H.; Wrana, J.L. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 2000, 6, 1365–1375. [Google Scholar] [CrossRef]
- Ebisawa, T.; Fukuchi, M.; Murakami, G.; Chiba, T.; Tanaka, K.; Imamura, T.; Miyazono, K. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 2001, 276, 12477–12480. [Google Scholar] [CrossRef] [Green Version]
- Murakami, G.; Watabe, T.; Takaoka, K.; Miyazono, K.; Imamura, T. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell 2003, 14, 2809–2817. [Google Scholar] [CrossRef]
- Horiki, M.; Imamura, T.; Okamoto, M.; Hayashi, M.; Murai, J.; Myoui, A.; Ochi, T.; Miyazono, K.; Yoshikawa, H.; Tsumaki, N. Smad6/Smurf1 overexpression in cartilage delays chondrocyte hypertrophy and causes dwarfism with osteopenia. J. Cell Biol. 2004, 165, 433–445. [Google Scholar] [CrossRef]
- Jia, J.; Feng, X.; Xu, W.; Yang, S.; Zhang, Q.; Liu, X.; Feng, Y.; Dai, Z. MiR-17-5p modulates osteoblastic differentiation and cell proliferation by targeting SMAD7 in non-traumatic osteonecrosis. Exp. Mol. Med. 2014, 46, e107. [Google Scholar] [CrossRef] [Green Version]
- Wei, B.; Wei, W.; Zhao, B.; Guo, X.; Liu, S. Long non-coding RNA HOTAIR inhibits miR-17-5p to regulate osteogenic differentiation and proliferation in non-traumatic osteonecrosis of femoral head. PLoS ONE 2017, 12, e0169097. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Xiang, Q.; Jia, J.; Guo, T.; Liao, Z.; Yang, S.; Cai, X.; Liu, X. CircHGF suppressed cell proliferation and osteogenic differentiation of BMSCs in ONFH via inhibiting miR-25-3p binding to SMAD7. Mol. Ther. Nucleic Acids 2022, 28, 99–113. [Google Scholar] [CrossRef]
- Han, N.; Qian, F.; Niu, X.; Chen, G. Circ_0058792 regulates osteogenic differentiation through miR-181a-5p/Smad7 axis in steroid-induced osteonecrosis of the femoral head. Bioengineered 2022, 13, 12807–12822. [Google Scholar] [CrossRef]
- Fang, S.H.; Chen, L.; Chen, H.H.; Li, Y.F.; Luo, H.B.; Hu, D.Q.; Chen, P. MiR-15b ameliorates SONFH by targeting Smad7 and inhibiting osteogenic differentiation of BMSCs. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 9761–9771. [Google Scholar] [CrossRef]
- Hao, Y.; Lu, C.; Zhang, B.; Xu, Z.; Guo, H.; Zhang, G. CircPVT1 up-regulation attenuates steroid-induced osteonecrosis of the femoral head through regulating miR-21-5p-mediated Smad7/TGFβ signalling pathway. J. Cell. Mol. Med. 2021, 25, 4608–4622. [Google Scholar] [CrossRef]
- Jiang, L.B.; Tian, L.; Zhang, C.G. Bone marrow stem cells-derived exosomes extracted from osteoporosis patients inhibit osteogenesis via microRNA-21/SMAD7. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6221–6229. [Google Scholar] [CrossRef] [PubMed]
- Selvamurugan, N.; He, Z.; Rifkin, D.; Dabovic, B.; Partridge, N.C. Pulsed electromagnetic field regulates microRNA 21 expression to activate TGF-β signaling in human bone marrow stromal cells to enhance osteoblast differentiation. Stem Cells Int. 2017, 2017, 2450327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valenti, M.T.; Deiana, M.; Cheri, S.; Dotta, M.; Zamboni, F.; Gabbiani, D.; Schena, F.; Dalle Carbonare, L.; Mottes, M. Physical exercise modulates miR-21-5p, miR-129-5p, miR-378-5p, and miR-188-5p expression in progenitor cells promoting osteogenesis. Cells 2019, 8, 742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Guo, L.; Liu, Y.; Su, Y.; Xie, Y.; Du, J.; Zhou, J.; Ding, G.; Wang, H.; Bai, Y.; et al. MicroRNA-21 promotes osteogenesis of bone marrow mesenchymal stem cells via the Smad7-Smad1/5/8-Runx2 pathway. Biochem. Biophys. Res. Commun. 2017, 493, 928–933. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Zhong, L.; Chen, C.; Tang, Z.; Liu, H.; Zhou, Y.; Tang, M.; Zhou, L.; Zuo, G.; Luo, J.; et al. MiR-21 synergizes with BMP9 in osteogenic differentiation by activating the BMP9/Smad signaling pathway in murine multilineage cells. Int. J. Mol. Med. 2015, 36, 1497–1506. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, B.; Balagangadharan, K.; Selvamurugan, N. Syringic acid, a phenolic acid, promotes osteoblast differentiation by stimulation of Runx2 expression and targeting of Smad7 by miR-21 in mouse mesenchymal stem cells. J. Cell Commun. Signal. 2018, 12, 561–573. [Google Scholar] [CrossRef]
- Sanjeev, G.; Sidharthan, D.S.; Pranavkrishna, S.; Pranavadithya, S.; Abhinandan, R.; Akshaya, R.L.; Balagangadharan, K.; Siddabathuni, N.; Srinivasan, S.; Selvamurugan, N. An osteoinductive effect of phytol on mouse mesenchymal stem cells (C3H10T1/2) towards osteoblasts. Bioorg. Med. Chem. Lett. 2020, 30, 127137. [Google Scholar] [CrossRef]
- Li, H.; Yang, F.; Wang, Z.; Fu, Q.; Liang, A. MicroRNA-21 promotes osteogenic differentiation by targeting small mothers against decapentaplegic 7. Mol. Med. Rep. 2015, 12, 1561–1567. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Jiang, D. Hypoxia-responsive miRNA-21-5p inhibits Runx2 suppression by targeting SMAD7 in MC3T3-E1 cells. J. Cell. Biochem. 2019, 120, 16867–16875. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.C.; Yang, M.H.; Tsai, C.C.; Huang, T.F.; Chen, Y.H.; Hung, S.C. Hypoxia inhibits osteogenesis in human mesenchymal stem cells through direct regulation of RUNX2 by TWIST. PLoS ONE 2011, 6, e23965. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Sun, C.; Lu, J.; Zou, L.; Hu, M.; Yang, Z.; Xu, Y. MicroRNA-590-5p antagonizes the inhibitory effect of high glucose on osteoblast differentiation by suppressing Smad7 in MC3T3-E1 cells. J. Int. Med. Res. 2019, 47, 1740–1748. [Google Scholar] [CrossRef] [Green Version]
- Vishal, M.; Vimalraj, S.; Ajeetha, R.; Gokulnath, M.; Keerthana, R.; He, Z.; Partridge, N.C.; Selvamurugan, N. MicroRNA-590-5p stabilizes Runx2 by targeting Smad7 during osteoblast differentiation. J. Cell. Physiol. 2017, 232, 371–380. [Google Scholar] [CrossRef]
- Liu, T.; Wu, Y.; Huang, T.; Zhang, X.; Cai, Y. MiR-590 promotes the proliferation of HUMSCs and induces ECM synthesis by targeting Smad7. Oncol. Lett. 2017, 14, 3941–3946. [Google Scholar] [CrossRef] [Green Version]
- He, G.; Chen, J.; Huang, D. MiR-877-3p promotes TGF-β1-induced osteoblast differentiation of MC3T3-E1 cells by targeting Smad7. Exp. Ther. Med. 2019, 18, 312–319. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.P.; Li, Q.W.; Shu, Z.Z.; Liu, Y. TP53-mediated miR-2861 promotes osteogenic differentiation of BMSCs by targeting Smad7. Mol. Cell. Biochem. 2022, 477, 283–293. [Google Scholar] [CrossRef]
- Xu, W.; Xia, R.; Tian, F.; Liu, L.; Li, M.; Fang, S. MicroRNA-324-3p promotes osteoblasts differentiation via suppressing SMAD7. J. Hard Tissue Biol. 2022, 31, 263–268. [Google Scholar] [CrossRef]
- Song, Y.; Mou, R.; Li, Y.; Yang, T. Zingerone promotes osteoblast differentiation via miR-200c-3p/smad7 regulatory axis in human bone mesenchymal stem cells. Med. Sci. Monit. 2020, 26, e919309. [Google Scholar] [CrossRef]
- Peng, Z.; Mai, Z.; Xiao, F.; Liu, G.; Wang, Y.; Xie, S.; Ai, H. MiR-20a: A mechanosensitive microRNA that regulates fluid shear stress-mediated osteogenic differentiation via the BMP2 signaling pathway by targeting BAMBI and SMAD6. Ann. Transl. Med. 2022, 10, 683. [Google Scholar] [CrossRef]
- Huang, W.; Wu, Y.; Zhao, Y.; Gan, X.; Zhang, B.; Cen, X.; Huang, X.; Zhao, Z. Down-regulation of hsa-circ-0107593 promotes osteogenic differentiation of hADSCs via miR-20a-5p/SMAD6 signaling. Oral Dis. 2022. Advance online publication. [Google Scholar] [CrossRef]
- Czekanska, E.M.; Stoddart, M.J.; Richards, R.G.; Hayes, J.S. In search of an osteoblast cell model for In Vitro research. Eur. Cells Mater. 2012, 24, 1–17. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. EJIFCC 2019, 30, 114–127. [Google Scholar] [PubMed]
- Chen, Y.; Zhao, H.; Tan, Z.; Zhang, C.; Fu, X. Bottleneck limitations for microRNA-based therapeutics from bench to the bedside. Die Pharm. 2015, 70, 147–154. [Google Scholar]
- Winkle, M.; El-Daly, S.M.; Fabbri, M.; Calin, G.A. Noncoding RNA therapeutics—Challenges and potential solutions. Nat. Rev. Drug Discov. 2021, 20, 629–651. [Google Scholar] [CrossRef] [PubMed]





| miRNAs | Protein Encoded by Direct Target Gene(s) | Model Systems and Cell Types | Gene Targeting Effect of miRNAs on Osteogenesis | References |
|---|---|---|---|---|
| MicroRNAs and Transforming Growth Factor-β (TGF-β) Ligands | ||||
| MiR-185 | TGF-β1 | Bone and peripheral blood samples obtained from patients with spinal cord injuries induced by thoracolumbar spine compression fractures | − | [103] |
| TGF-β1 | Bone and peripheral blood samples obtained from male and female patients of ankle fracture (23–60 years old); hFOB1.19 cells | − | [104] | |
| MiR-675 | TGF-β1 | Primary human MSCs; osteogenically induced human MSCs implanted subcutaneously in BALB/c homozygous nude (nu/nu) mice (5 weeks old) | + | [110] |
| MiR-141 | TGF-β2 | Male and female SONFH patients (mean age: 60.45 years) and control patients with femoral neck fractures (mean age: 60.85 years); SONFH male and female Sprague–Dawley (SD) rat model (body weight 100 ± 20 g) | − | [107] |
| MiR-29b | TGF-β3, HDAC4, ACVR2A, CTNNBIP1, and DUSP2; COL1A1, COL5A3, COL4A2 | MC3T3-E1 clone 4 cells; primary rat calvaria osteoblast cells | −/+ | [111] |
| MiR-140-3p | TGF-β3 | MC3T3-E1 cells | −/+ | [109] |
| MicroRNAs and TGF-β Receptors (TGFBR) | ||||
| MiR-140-5p | TGFβRI | db/db obese mice and their genetically matched lean littermates (10 weeks old); BMSCs isolated from femurs and tibias of C57 mice (4 weeks old); stromal ST2 and preadipocyte 3T3-L1 cells | − | [123,125] |
| Let-7a-5p | TGFβRI | BMSCs derived from OVX female C57BL/6J mice (4 weeks old, body weight 20 ± 2 g); human embryonic kidney HEK cells | − | [117] |
| Let-7f-5p | TGFβRI | BMSCs derived bone marrow collected from long bones (tibias and femurs) of mice (8 weeks old); glucocorticoid-induced osteoporosis model of male C57BL/6 mice (8 weeks old) | + | [120] |
| MiR-142-3p | TGFβRI | Male C57BL/6 mice fracture model and traumatic brain injury model (5–6 weeks old); regulatory T cells culture; HUVECs; mouse BMSCs isolated from fresh femoral bone marrow samples; isolation and treatment of TregD-Exos | + | [121] |
| MiR-181 isoforms (miR-181a, miR-181b, and miR-181c) | TGFβRI, Tgfbi | C2C12 cells; MC3T3-E1 cells; primary calvaria osteoblasts; tibia and calvaria samples harvested from male new-born (24 h) C57BL/6 wildtype mice (1–6 weeks old) | + | [122] |
| MiR-223 | TGFBRII, FGFR2 | Periodontitis patients and healthy controls; periodontal ligament tissues collected from 30 teeth, which were premolars and third molars extracted from healthy donors (14–23 years old) | − | [124] |
| miRNAs | Protein Encoded by Direct Target Gene(s) | Model Systems and Cell Types | Gene Targeting Effect of miRNAs on Osteogenesis | References |
|---|---|---|---|---|
| MicroRNAs and Bone Morphogenic Protein (BMP) Ligands | ||||
| MiR-20a | PPARγ, Bambi, Crim1 | Human BMSCs from healthy donors | + | [140] |
| MiR-20a-5p | Bambi | Human DPSCs isolated from premolars collected from donors who underwent teeth extraction Due to orthodontic needs (12–14 years old); female nude BALB/c mice with calvarial defects (32 months old) | + | [143] |
| MiR-140-5p and miR-140-3p | BMP-2 | Human MSCs | − | [108] |
| MiR-140-5p | BMP-2 | Mouse BMSCs | − | [158] |
| MiR-204 | BMP-2 | BMSCs isolated from male Sprague–Dawley (SD) rats (4 weeks old) | − | [144] |
| MiR-98 (let-7/miR-98 family) | BMP-2 | BMSCs isolated from femoral head of male and female patients undergoing hip arthroplasty (mean age: 59.1 ± 6.2 years) | − | [145] |
| MiR-98-5p | BMP-2 | High glucose-treated MC3T3-E1 cells | − | [146,139] |
| MiR-214 | BMP-2 | OVX female Sprague–Dawley (SD) rats (12 weeks old, body weight 250–300 g) which underwent subsequent osteoporotic fracture operation | − | [147] |
| BMP-2 | Fibroblasts isolated from the cut hip joint capsule ligament of male and female patients with AS Involving both hips and requiring joint replacement (average age 40.7 ± 2.4 years), and control patients with femoral neck fracture (free of AS and other immune diseases) who needed open surgery or joint replacement (average age 40.7 ± 2.4 years) | − | [150] | |
| BMP-2 | Human BMSCs | − | [157] | |
| MiR-410 | BMP-2 | Female patients with postmenopausal osteoporosis (age range 50–59 years old, mean age: 55.6 ± 4.8 years) and healthy female subjects (age range 50–59 years old, mean age: 55.1 ± 4.6 years); female C57BL/6 mice (5 weeks old, weight between 18–22 g); CD14+ peripheral blood mononuclear cells (PBMCs) | − | [148] |
| MiR-93-5p | BMP-2 | Male and female patients with femoral neck fractures; human BMSCs isolated from the bone marrow of donors with trauma | − | [151] |
| MiR-543 | BMP-2 | MC3T3-E1 cells | − | [154] |
| MiR-142 | BMP-2 | MC3T3-E1 subclone 14 cells | − | [155] |
| MiR-342-5p | COL4A6, BMP-2 | MC3T3-E1 cells, human MSCs procured from bone marrow aspirate and hMSC-TERT cell line; male and female fracture patients including hand fracture (25–56 years old), intra-articular calcaneal fracture (24–59 years old), healthy controls (24–59 years old) | − | [156] |
| MiR-6979-5p | BMP-2 | Male C57BL/6J mice fracture modal (6 weeks old); MC3T3-E1 cells | − | [159] |
| MiR-1323 | BMP-4 | Human male atrophic non-union fracture specimens and standard healing fracture specimens collected during open reduction/internal fixation (ORIF); MSCs; male Sprague–Dawley (SD) rat model of femoral fracture (13–14 weeks old, body weight 400–500 g) | − | [182] |
| MiR-451a | BMP-6 | Bone samples of wild-type and miR-451a-knockout (KO) mice with or without OVX (6 weeks old), and miR-451a overexpression mice following OVX; primary osteoblasts isolated from neonatal mice (3 days old); mouse BMSCs obtained from femurs and tibias | − | [160] |
| MiR-765 | BMP-6 | Human MSCs | − | [161] |
| MiR-146a-5p and miR-146b-5p | BMP-6 | Human PDLSCs isolated from normal impacted third molars collected from human subjects (16–35 years old) | − | [162] |
| MiR-542-3p | BMP-7 | Mouse calvarial osteoblasts; OVX female BALB/c mice (6 weeks old) | − | [163] |
| MiR-485-5p | BMP-7 | Bone marrow samples and primary human BMSCs harvested from osteoporosis patients and healthy controls | − | [164] |
| MiR-181a-3p | BMP-10 | Human BMSCs obtained from healthy volunteers | − | [165] |
| MiR-450b | BMP-3 | Human adipose tissue obtained from donors (20–45 years old) undergoing liposuction; scaffolds loaded with human ADSCs infected with lenti-450b implanted subcutaneously into the upper dorsal surface of male NOD/SCID mice (6 weeks old) | + | [166] |
| MiR-1273g-3p | BMP-3 | Human breast cancer cells, MCF-7, HS598t, and MDA-MB-231; human normal mammary epithelial cells, MCF-10A; co-culture of shSNHG3 transfected MD-MB-231 with human BMSCs; SNHG3 loss-of-function analysis in female C57BL/6 mice which underwent segmental defect operation of femur (2 months old) | + | [167] |
| MicroRNAs and Bone Morphogenic Protein Receptor (BMPR) | ||||
| MiR-100 | BMPR2 | Human ADSCs isolated from adipose tissues obtained from patients undergoing tumescent liposuction | − | [173] |
| MiR-153 | BMPR2 | Human BMSCs isolated from bone marrow samples of young subjects (<30 years old) and older subjects (>60 years old) with slight or severe osteoporosis | − | [174] |
| MiR-155 | BMPR2 | HEK293 human embryonic kidney cell lines; human C2C12 myoblast cell lines; mouse embryonic fibroblasts (MEF); ectopic in vivo bone formation assay of transfected MEF cells in athymic female nude mice (4–6 weeks old) | − | [175] |
| MiR-494 | BMPR2 | C2C12 and HEK293T cells; osteoblasts isolated from tail-suspended rats mimics a simulated microgravity environment | − | [176] |
| MiR-1187 | BMPR2, ArhGEF-9 | Primary calvarial osteoblasts harvested from mouse pups (1–2 days old); miR-1187 gain-of-function and loss-of-function analysis in BALB/c neonatal pups (1–2 days old) and post-OVX female BALB/c mice (6 weeks old) | − | [177] |
| MiR-195-5p | BMPR1A, Wnt3a, FGF2 | Human PDL tissues obtained from premolars that previously extracted from healthy individuals (14–20 years old) for orthodontic purposes; PDL cells subjected to cyclic tension stress (CTS) for modelling of orthodontic mechanical loading in vitro; C57BL/6 mouse model of mechanical loading tooth movement | − | [178] |
| MiR-23a | BMPR1B | Patients with moderate to advanced periodontitis (18–60 years old) and healthy subjects; gingival crevicular fluid samples; human PDLSCs isolated from tissues in the middle of teeth roots | − | [179] |
| MiR-125b | BMPR1B | Human BMSCs isolated from the posterior iliac crest of young healthy male volunteers; male BALB/c nude mice model of bone defects (7 weeks old, body weight 16–23.5 g) | − | [180] |
| MiR-497∼195 cluster (miR-15 family) | BMP-responsive genes, such as Furin, Acvr2a, Bmpr1a, Dies1, Tgfbr3, Smad5, Ski, Zfp423, Mapk3, and Smurf1 | Bone tissue samples harvested from C57BL/6 wild-type mice; primary calvaria osteoblasts harvested from new-born mice (P0-P4); MC3T3-E1 subclone 4 cells | −/+ | [139] |
| miRNAs | Protein Encoded by Direct Target Gene(s) | Model Systems and Cell Types | Gene Targeting Effect of miRNAs on Osteogenesis | References |
|---|---|---|---|---|
| MicroRNAs and the BMP-Regulated Smads (R-Smads)—Smad1 and Smad5 | ||||
| MiR-26a | Smad1 | Human ADSCs isolated from adipose tissue obtained from the subcutaneous abdominal depot during herniotomy | − | [191] |
| Smad1 | C57BL/6J mice (4–6 weeks old); mouse BMSCs isolated from the bone marrow cavities of femur and tibia; mouse ADSCs isolated from scraps of subcutaneous adipose tissues | − | [192] | |
| MiR-30 family (miR-30a, miR-30b, miR-30c, and miR-30d) | Smad1 | MC3T3-E1 cells; mouse BMSCs prepared from the bone marrow of femur and tibia harvested from male C57B/L6 mice (2 months old) | − | [194] |
| MiR-100 | Smad1 | MC3T3-E1 cells; mouse BMSCs isolated and harvested from femur and tibia bone marrow of male C57B/L6 mice (10 weeks old) | − | [197] |
| MiR-203-3p | Smad1 | Diabetic male Sprague–Dawley (SD) rats (10 weeks old, body weight 290–310 g); high-glucose treated rat BMSCs isolated from mandible of male SD rats (8 weeks old) and C3H101/2 clone 8 cells | − | [201] |
| MiR-222-3p | Smad5 | Human foetal MSCs | – | [202] |
| MiR-128-3p | Smad5 | Male femoral fracture Sprague–Dawley (SD) rats (3 months old); BMSCs obtained from young (4 weeks old) and old (72 weeks old) male SD rats | − | [203] |
| MiR-320a | Smad5 | Human BMSCs isolated from the bone marrow of healthy subjects | − | [204] |
| MiR-133 and miR-135 | Smad5 | C2C12 cells | − | [193] |
| MiR-133a, miR-133b, and miR-135 | Smad5 | Human DPSCs cultured on titanium disks in vitro | − | [205] |
| MiR-24-3p | Smad5 | PDLSCs isolated from human first premolars extracted for orthodontic reasons from donors (14–20 years old) | − | [207] |
| MiR-24-1-5p | Smad5 | Female C57BL/6 mice (8–10 weeks old); mouse offspring; primary mouse calvarial osteoblasts obtained from new-born pups of the control and high-protein group | − | [211] |
| MiR-21 | Smad5 | PDLSCs isolated from human first premolars extracted for orthodontic reasons from donors (10–14 years old); miR-21 gain-of-function and loss-of-function analysis of transfected PDLSCs implanted on immunocompromised beige mice (nu/nu nude mice) (10 weeks old) | − | [206] |
| MiR-106b-5p/miR-17-5p | Smad5 | C2C12 cells; MC3T3-E1 cells; OVX female C57BL/6J mice (6 weeks old) | − | [208] |
| MiR-155 | Smad5 | MC3T3-E1 cells and HEK293 cells | − | [209] |
| MiR-132-3p | Smad5 | MC3T3-E1 cells loaded with cyclic stress | − | [210] |
| MicroRNAs and the TGF-β-Regulated Smads (R-Smads)—Smad2 and Smad3 | ||||
| MiR-25-5p | Smad2 | BMSCs isolated from Sprague Dawley rats (4 weeks old) and infected with an adenovirus vector encoding the full-length sequence of Rattus norvegicus Nell1, followed by Nell1/EVs isolation; in vivo evaluation of bone repair with the Nell1/EV-hydrogel conducted on calvarial defect male SD rat model (6 weeks old) | − | [214] |
| MiR-10b | Smad2 | Human ADSCs isolated from adipose tissue collected from healthy women who underwent Liposuction surgery; osteoporosis patients who had a fracture caused by falling without obvious violence; miR-10b gain-of-function and ectopic bone formation model of NOD/SCID mice (8 weeks old) | + | [215] |
| MiR-214-5p | TGF-β, Smad2, COL44A1 | PTA-1058 human BMSC cell line | − | [216] |
| MiR-6315 | Smad2 | MC3T3-E1 pre-osteoblastic cells and 3T3 F442A pre-adipocytic cells | + | [217] |
| MiR-146a | Smad2, Smad3 | Osteogenic diaphyseal and chondrogenic epiphyseal foetal femur cells population isolated from foetal femurs samples at 7–9 weeks post conception | + | [220] |
| MiR-708 | Smad3 | Bone marrow samples of patients with SONFH and patients with ONFH after a previous fracture of the femoral neck (both ranged 20–50 years old); human BMSCs isolated from the proximal end of femur after inserting the tapered awl into the femoral canal during THA; glucocorticoid-treated MSCs | − | [224] |
| MiR-181d | Smad3 | Whole-bone marrow samples from the marrow cavity of patients undergoing total hip arthroplasty due to steroid-induced necrosis of the femoral head and control patients with secondary femoral head necrosis after the old femoral neck fracture (both ranged 20–50 years old); human BMSCs isolated from the proximal end of femur | − | [225] |
| MiR-596 | Smad3 | Bone marrow samples from patients with SONFH and from patients with femoral neck fracture who underwent total hip replacement (both ranged 25–50 years old); glucocorticoid-treated BMSCs | − | [226] |
| MiR-423-5p | Smad3 | Serum samples from SONFH patients (50.5 ± 2.7 years old, 45% male) and healthy volunteers (49.5 ± 3.1 years old, 55% male) | − | [227] |
| Smad3 | Bone marrow samples from non-traumatic ONFH patients and OA control patients; human BMSCs and HEK293T cells | − | [228] | |
| MiR-23b | Smad3 | Lipopolysaccharides-treated MC3T3-E1 cells | + | [231] |
| MiR-300 | Smad3 | Primary rat osteoblast cells isolated from neonatal rat pups; human osteoblasts; femur and tibia collected from PepC-treated Wistar rats (3 months old); miR-300 gain-of-function and loss-of- function analysis in rat pups (2–3 days old); miR-300 gain-of-function and loss-of-function analysis in OVX Wistar albino rats (3 months old, body weight 180 g); blood serums from postmenopausal osteoporotic and non-osteoporotic human subjects | − | [232] |
| MicroRNAs and the Partner or Common Smad (Co-Smad)—Smad4 | ||||
| MiR-224 | Smad4 | MSC cell lines, including MSC derived from bone marrow (MSC-B), MSC umbilical cord-derived (MSC-U), MSC adipose-derived (MSC-A), and human skull osteoblasts (HCO); HEK293T cells | − | [234] |
| MiR-224-5p | Smad4 | BMSCs extracted from the tibias and femurs of Sprague–Dawley (SD) rats (6 weeks old); SONFH male Sprague–Dawley SD rat model (12 weeks old) | − | [235] |
| MiR-1323 | Smad4 | Human male atrophic non-union fracture specimens and standard healing fracture specimens collected during open reduction/internal fixation (ORIF); MSCs; male Sprague–Dawley (SD) rat model of femoral fracture (13–14 weeks old, body weight 400–450 g) | − | [182] |
| MiR-146a | Smad4 | BMSCs isolated from male C57Bl6 mice (8-weeks old) | − | [237] |
| MiR-144-3p | Smad4 | C3H10T1/2 cells | − | [238] |
| MiR-664-3p | Smad4, osterix | MC3T3-E1 cells; HEK293T cells; C3H10T1/2 cells; bone tissues of C57BL/6J mice (8 weeks old); conditional miR-664-3p transgenic C57BL/6J mice; miR-664-3p loss-of-function analysis in post- operated OVX female C57BL/6J mice (16 weeks old); peripheral blood samples of osteoporotic female patients with fracture and non-osteoporotic patients (50–89 years old) | − | [239] |
| MicroRNAs and the Inhibitory Smads (I-Smads)—Samd6 and Smad7 | ||||
| MiR-17-5p | Smad7 | Bone marrow specimens obtained from the proximal end of the femurs derived from non- traumatic ONFH patients and OA control patients; primary human BMSCs derived from the patients; commercial hMSC-BM cells | + | [247] |
| Smad7 | Bone marrow specimens obtained from the proximal end of the femurs derived from non- traumatic ONFH patients and OA control patients; primary human BMSCs derived from the patients; commercial hMSC-BM cells | + | [248] | |
| MiR-25-3p | Smad7 | Bone marrow specimens were harvested from patients with steroid-induced ONFH (25–68 years old, mean 50.0 years) and control patients with femoral neck fracture (33–68 years old, mean 54.6 years); human BMSCs | + | [249] |
| MiR-181a-3p | Smad7 | BMSCs derived from patients with steroid-induced ONFH; methylprednisolone-induced ONFH Sprague–Dawley (SD) female rats (body weight 190–220 g); MC3T3-E1 cells | + | [250] |
| MiR-15b | Smad7 | Bone marrow tissues collected from patients with glucocorticoid-induced osteonecrosis of the femoral head and patients with secondary ONFH receiving total hip replacement (both ranged 25–50 years old); human BMSCs; HEK293T cells | + | [251] |
| MiR-21 | Smad7 | MSCs extracted from osteoporosis patients and healthy adults | − | [253] |
| Smad7 | BMSCs extracted from fresh human bone marrows of ‘young’ female patients (21–30 years old) and ‘older’ female patients (31–65 years old) which treated with PEMF | + | [254] | |
| Smad7 | BMSCs isolated from the femur and tibia bone cavities of female C57BL/6J wild-type mice and miR-21-knockout (KO) mice (5 weeks old); wild-type and miR-21-KO mouse model of calvarial bone defects (body weight 20–22 g) | + | [256] | |
| Smad7 | Human HEK293 cells, colorectal carcinoma HCT116 cells, and murine multilineage cells (MMCs): murine C2C12 myoblasts cell lines, and mouse embryonic fibroblasts (MEFs) | + | [257] | |
| Smad7 | Syringic acid-treated mouse MSCs | + | [258] | |
| Smad7 | MC3T3-E1 cells | + | [260] | |
| MiR-21-5p, miR-129-5p, miR-378-5p | PTEN, Smad7 | Serum samples collected before and immediately after a 21.1 km half marathon from male amateur runners (median age 40.2 ± 8 years); human BMSCs | + | [255] |
| MiR-21-5p | Smad7 | SONFH male Sprague–Dawley (SD) rat model and control groups (10 weeks old); rat BMSCs isolated from bone marrow tissues removed from the proximal femur during total hip replacement | − | [252] |
| Smad7 | Hypoxia-treated MC3T3-E1 cells | + | [261] | |
| MiR-21a | Smad7 | Phytol-treated C3H10T1/2 cells | + | [259] |
| MiR-590-5p | Smad7 | High glucose-treated MC3T3-E1 cells | + | [263] |
| MiR-590-5p | Smad7 | Human BMSCs obtained from female subjects aged 27, 29, 30, and 36 years; mouse MSC C3H10T1/2 cells and human MG-63 cells | + | [264] |
| MiR-590 | Smad7 | Human UMSCs | + | [265] |
| MiR-877-3p | Smad7 | MC3T3-E1 cells; miR-877-3p gain-of-function and loss-of-function analysis in female NOD/SCID mice (4 weeks old, body weight 17–19 g) | + | [266] |
| MiR-324-3p | Smad7 | Icarrin-treated MC3T3-E1 cells | + | [268] |
| MiR-200c | Smad7 | Zingerone-treated human BMSCs | + | [269] |
| MiR-20a | Smad6, Bambi | Fluid sheer stress-treated MC3T3-E1 cells | + | [270] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loh, H.-Y.; Norman, B.P.; Lai, K.-S.; Cheng, W.-H.; Nik Abd. Rahman, N.M.A.; Mohamed Alitheen, N.B.; Osman, M.A. Post-Transcriptional Regulatory Crosstalk between MicroRNAs and Canonical TGF-β/BMP Signalling Cascades on Osteoblast Lineage: A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 6423. https://doi.org/10.3390/ijms24076423
Loh H-Y, Norman BP, Lai K-S, Cheng W-H, Nik Abd. Rahman NMA, Mohamed Alitheen NB, Osman MA. Post-Transcriptional Regulatory Crosstalk between MicroRNAs and Canonical TGF-β/BMP Signalling Cascades on Osteoblast Lineage: A Comprehensive Review. International Journal of Molecular Sciences. 2023; 24(7):6423. https://doi.org/10.3390/ijms24076423
Chicago/Turabian StyleLoh, Hui-Yi, Brendan P. Norman, Kok-Song Lai, Wan-Hee Cheng, Nik Mohd Afizan Nik Abd. Rahman, Noorjahan Banu Mohamed Alitheen, and Mohd Azuraidi Osman. 2023. "Post-Transcriptional Regulatory Crosstalk between MicroRNAs and Canonical TGF-β/BMP Signalling Cascades on Osteoblast Lineage: A Comprehensive Review" International Journal of Molecular Sciences 24, no. 7: 6423. https://doi.org/10.3390/ijms24076423