Crystal Structure of Pyrrolysyl-tRNA Synthetase from a Methanogenic Archaeon ISO4-G1 and Its Structure-Based Engineering for Highly-Productive Cell-Free Genetic Code Expansion with Non-Canonical Amino Acids
Abstract
:1. Introduction
2. Results
2.1. Overall Structure of ISO4-G1 PylRS
2.2. Structure-Based Sequence Comparison of ISO4-G1 PylRS with Other PylRSs
2.3. Structural Comparison of ISO4-G1 PylRS with MaPylRS, DhPylSc, and MmPylRSc
2.4. Structural Comparison of the Amino Acid Binding Residues of ISO4-G1 PylRS with Those of MaPylRS and MmPylRSc
2.5. Structural Changes of Tyr204 and His225 in ISO4-G1 PylRS and Comparison with Those of MmPylRS and MaPylRS
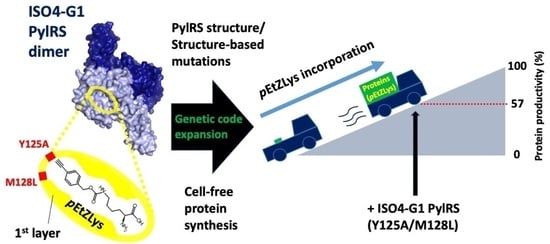
2.6. Structure-Based Engineering of the First-Layer Residues in ISO4-G1 PylRS for Site-Specific Incorporation of Bulky Lysine Derivatives into Proteins by Cell-Free Protein Synthesis
2.7. Effects of the ISO4-G1 PylRS(Y125A/M128L) Concentration on Cell-Free Protein Synthesis with the Inefficient Amino Acid pEtZLys
2.8. Effects of the Second-Layer Mutations of ISO4-G1 PylRS for Site-Specific Incorporation of Bulky Lysine Derivatives into Proteins by Cell-Free Protein Synthesis
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Bacterial Strains and Plasmids
4.3. Expression and Purification of PylRS Proteins
4.4. Preparation of tRNAPyl Transcripts
4.5. Crystallization, Data Collection, and Structure Determination
4.6. Cell-Free Protein Synthesis and Purification of GFP Proteins Containing Non-Canonical Amino Acids
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Wang, L.; Xie, J.; Schultz, P.G. Expanding the genetic code. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.C.; Schultz, P.G. Adding New Chemistries to the Genetic Code. Annu. Rev. Biochem. 2010, 79, 413–444. [Google Scholar] [CrossRef] [Green Version]
- Blight, S.K.; Larue, R.C.; Mahapatra, A.; Longstaff, D.G.; Chang, E.; Zhao, G.; Kang, P.T.; Green-Church, K.B.; Chan, M.K.; Krzycki, J.A. Direct charging of tRNACUA with pyrrolysine in vitro and in vivo. Nature 2004, 17, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Polycarpo, C.; Ambrogelly, A.; Bérubé, A.; Winbush, S.A.M.; McCloskey, J.A.; Crain, P.F.; Wood, J.L.; Söll, D. An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. USA 2004, 101, 12450–12454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrogelly, A.; Gundllapalli, S.; Herring, S.; Polycarpo, C.; Frauer, C.; Söll, D. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proc. Natl. Acad. Sci. USA 2007, 104, 3141–3146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, H.; Peak-Chew, S.Y.; Chin, J.W. Genetically encoding Nε-acetyllysine in recombinant proteins. Nat. Chem. Biol. 2008, 4, 232–234. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Kobayashi, T.; Hino, N.; Yanagisawa, T.; Sakamoto, K.; Yokoyama, S. Adding L-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 2008, 371, 818–822. [Google Scholar] [CrossRef]
- Yanagisawa, T.; Ishii, R.; Fukunaga, R.; Kobayashi, T.; Sakamoto, K.; Yokoyama, S. Multistep Engineering of Pyrrolysyl-tRNA Synthetase to Genetically Encode Nε-(o-Azidobenzyloxycarbonyl) lysine for Site-Specific Protein Modification. Chem. Biol. 2008, 15, 1187–1197. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.R.; Groff, D.; Guo, J.; Ou, W.; Cellitti, S.; Geierstanger, B.H.; Schultz, P.G. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chemie-Int. Ed. 2009, 48, 4052–4055. [Google Scholar] [CrossRef]
- Wan, W.; Tharp, J.M.; Liu, W.R. Pyrrolysyl-tRNA synthetase: An ordinary enzyme but an outstanding genetic code expansion tool. Biochim. Biophys. Acta-Proteins Proteom. 2014, 1844, 1059–1070. [Google Scholar] [CrossRef] [Green Version]
- Hao, B.; Gong, W.; Ferguson, T.K.; James, C.M.; Krzycki, J.A.; Chan, M.K. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science 2002, 296, 1462–1466. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, G.; James, C.M.; Krzycki, J.A. Pyrrolysine encoded by UAG in archaea: Charging of a UAG-decoding specialized tRNA. Science 2002, 296, 1459–1462. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.M.; Jiang, R.; Jain, R.; Larue, R.C.; Krzycki, J.; Chan, M.K. Structure of Desulfitobacterium hafniense PylSc, a pyrrolysyl-tRNA synthetase. Biochem. Biophys. Res. Commun. 2008, 374, 470–474. [Google Scholar] [CrossRef]
- Nozawa, K.; O′Donoghue, P.; Gundllapalli, S.; Araiso, Y.; Ishitani, R.; Umehara, T.; Söll, D.; Nureki, O. Pyrrolysyl-tRNA synthetase-tRNAPyl structure reveals the molecular basis of orthogonality. Nature 2009, 457, 1163–1167. [Google Scholar] [CrossRef] [Green Version]
- Chin, J.W. Expanding and Reprogramming the Genetic Code of Cells and Animals. Annu. Rev. Biochem. 2014, 83, 379–408. [Google Scholar] [CrossRef]
- Crnković, A.; Suzuki, T.; Söll, D.; Reynolds, N.M. Pyrrolysyl-tRNA synthetase, an aminoacyl-tRNA synthetase for genetic code expansion. Croat. Chem. Acta 2016, 89, 163–174. [Google Scholar] [CrossRef]
- Brabham, R.; Fascione, M.A. Pyrrolysine Amber Stop-Codon Suppression: Development and Applications. ChemBioChem 2017, 18, 1973–1983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, J.W. Expanding and reprogramming the genetic code. Nature 2017, 550, 53–60. [Google Scholar] [CrossRef]
- Wang, L. Engineering the Genetic Code in Cells and Animals: Biological Considerations and Impacts. Acc. Chem. Res. 2017, 50, 2767–2776. [Google Scholar] [CrossRef]
- Vargas-Rodriguez, O.; Sevostyanova, A.; Söll, D.; Crnković, A. Upgrading aminoacyl-tRNA synthetases for genetic code expansion. Curr. Opin. Chem. Biol. 2018, 46, 115–122. [Google Scholar] [CrossRef]
- Tharp, J.M.; Ehnbom, A.; Liu, W.R. tRNAPyl: Structure, function, and applications. RNA Biol. 2018, 15, 441–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willis, J.C.W.; Chin, J.W. Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs. Nat. Chem. 2018, 10, 831–837. [Google Scholar] [CrossRef] [PubMed]
- Meineke, B.; Heimgärtner, J.; Lafranchi, L.; Elsässer, S.J. Methanomethylophilus alvus Mx1201 Provides Basis for Mutual Orthogonal Pyrrolysyl tRNA/Aminoacyl-tRNA Synthetase Pairs in Mammalian Cells. ACS Chem. Biol. 2018, 13, 3087–3096. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, A.; Iraha, F.; Ohtake, K.; Sakamoto, K. Pyrrolysyl-tRNA synthetase with a unique architecture enhances the availability of lysine derivatives in synthetic genetic codes. Molecules 2018, 23, 2460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beránek, V.; Willis, J.C.W.; Chin, J.W. An Evolved Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetase/tRNA Pair Is Highly Active and Orthogonal in Mammalian Cells. Biochemistry 2019, 58, 387–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seki, E.; Yanagisawa, T.; Kuratani, M.; Sakamoto, K.; Yokoyama, S. Fully Productive Cell-Free Genetic Code Expansion by Structure-Based Engineering of Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetase. ACS Synth. Biol. 2020, 9, 718–732. [Google Scholar] [CrossRef] [PubMed]
- Meineke, B.; Heimgärtner, J.; Eirich, J.; Landreh, M.; Elsässer, S.J. Site-Specific Incorporation of Two ncAAs for Two-Color Bioorthogonal Labeling and Crosslinking of Proteins on Live Mammalian Cells. Cell Rep. 2020, 31, 107811. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, E.H.; Qianzhu, H.; Tan, Y.J.; Adams, L.A.; Huber, T.; Otting, G. Genetic Encoding of N6-(((Trimethylsilyl)methoxy)carbonyl)-L-lysine for NMR Studies of Protein-Protein and Protein-Ligand Interactions. J. Am. Chem. Soc. 2021, 143, 1133–1143. [Google Scholar] [CrossRef]
- Cao, L.; Liu, J.; Ghelichkhani, F.; Rozovsky, S.; Wang, L. Genetic Incorporation of ϵ-N-Benzoyllysine by Engineering Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetase. Chembiochem 2021, 22, 2530–2534. [Google Scholar] [CrossRef]
- Fischer, J.T.; Söll, D.; Tharp, J.M. Directed Evolution of Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetase Generates a Hyperactive and Highly Selective Variant. Front. Mol. Biosci. 2022, 9, 850613. [Google Scholar] [CrossRef]
- Stieglitz, J.T.; Lahiri, P.; Stout, M.I.; Van Deventer, J.A. Exploration of Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetase Activity in Yeast. ACS Synth. Biol. 2022, 11, 1824–1834. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, E.H.; Qianzhu, H.; George, J.; Frkic, R.L.; Jackson, C.J.; Nitsche, C.; Otting, G.; Huber, T. Genetic Encoding of Cyanopyridylalanine for In-Cell Protein Macrocyclization by the Nitrile-Aminothiol Click Reaction. Angew. Chem. Int. Ed. Engl. 2022, 61, e202114154. [Google Scholar] [CrossRef] [PubMed]
- Avila-Crump, S.; Hemshorn, M.L.; Jones, C.M.; Mbengi, L.; Meyer, K.; Griffis, J.A.; Jana, S.; Petrina, G.E.; Pagar, V.V.; Karplus, P.A.; et al. Generating Efficient Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetases for Structurally Diverse Non-Canonical Amino Acids. ACS Chem. Biol. 2022, 17, 3458–3469. [Google Scholar] [CrossRef]
- Yanagisawa, T.; Umehara, T.; Sakamoto, K.; Yokoyama, S. Expanded genetic code technologies for incorporating modified lysine at multiple sites. ChemBioChem 2014, 15, 2181–2187. [Google Scholar] [CrossRef]
- Mukai, T.; Yanagisawa, T.; Ohtake, K.; Wakamori, M.; Adachi, J.; Hino, N.; Sato, A.; Kobayashi, T.; Hayashi, A.; Shirouzu, M.; et al. Genetic-code evolution for protein synthesis with non-natural amino acids. Biochem. Biophys. Res. Commun. 2011, 411, 757–761. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, T.; Takahashi, M.; Mukai, T.; Sato, S.; Wakamori, M.; Shirouzu, M.; Sakamoto, K.; Umehara, T.; Yokoyama, S. Multiple site-specific installations of Nε-monomethyl-L-lysine into histone proteins by cell-based and cell-free protein synthesis. ChemBioChem 2014, 15, 1830–1838. [Google Scholar] [CrossRef] [PubMed]
- Chemla, Y.; Ozer, E.; Schlesinger, O.; Noireaux, V.; Alfonta, L. Genetically expanded cell-free protein synthesis using endogenous pyrrolysyl orthogonal translation system. Biotechnol. Bioeng. 2015, 112, 1663–1672. [Google Scholar] [CrossRef] [PubMed]
- Seki, E.; Yanagisawa, T.; Yokoyama, S. Cell-free protein synthesis for multiple site-specific incorporation of noncanonical amino acids using cell extracts from RF-1 deletion E. coli strains. Methods Mol. Biol. 2018, 1728, 49–65. [Google Scholar] [CrossRef]
- Adachi, J.; Katsura, K.; Seki, E.; Takemoto, C.; Shirouzu, M.; Terada, T.; Mukai, T.; Sakamoto, K.; Yokoyama, S. Cell-free protein synthesis using S30 extracts from Escherichia coli RFzero strains for efficient incorporation of non-natural amino acids into proteins. Int. J. Mol. Sci. 2019, 20, 429. [Google Scholar] [CrossRef] [Green Version]
- Gerrits, M.; Budisa, N.; Merk, H. Site-Specific Chemoselective Pyrrolysine Analogues Incorporation Using the Cell-Free Protein Synthesis System. ACS Synth. Biol. 2019, 8, 381–390. [Google Scholar] [CrossRef]
- Chemla, Y.; Ozer, E.; Shaferman, M.; Zaad, B.; Dandela, R.; Alfonta, L. Simplified methodology for a modular and genetically expanded protein synthesis in cell-free systems. Synth. Syst. Biotechnol. 2019, 4, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Charna, R.A.; Des Soye, B.J.; Ntai, I.; Kelleher, N.L.; Jewett, M.C. An efficient cell-free protein synthesis platform for producing proteins with pyrrolysine-based noncanonical amino acids. Biotechnol. J. 2022, 17, e2200096. [Google Scholar] [CrossRef]
- Mukai, T.; Hayashi, A.; Iraha, F.; Sato, A.; Ohtake, K.; Yokoyama, S.; Sakamoto, K. Codon reassignment in the Escherichia coli genetic code. Nucleic Acids Res. 2010, 38, 8188–8195. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.B.F.; Xu, J.; Shen, Z.; Takimoto, J.K.; Schultz, M.D.; Schmitz, R.J.; Xiang, Z.; Ecker, J.R.; Briggs, S.P.; Wang, L. RF1 knockout allows ribosomal incorporation of unnatural amino acids at multiple sites. Nat. Chem. Biol. 2011, 7, 779–786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lajoie, M.J.; Rovner, A.J.; Goodman, D.B.; Aerni, H.R.; Haimovich, A.D.; Kuznetsov, G.; Mercer, J.A.; Wang, H.H.; Carr, P.A.; Mosberg, J.A.; et al. Genomically recoded organisms expand biological functions. Science 2013, 342, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.H.; Ntai, I.; Haimovich, A.D.; Kelleher, N.L.; Isaacs, F.J.; Jewett, M.C. Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth. Biol. 2014, 3, 398–409. [Google Scholar] [CrossRef]
- Mukai, T.; Hoshi, H.; Ohtake, K.; Takahashi, M.; Yamaguchi, A.; Hayashi, A.; Yokoyama, S.; Sakamoto, K. Highly reproductive Escherichia coli cells with no specific assignment to the UAG codon. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fredens, J.; Wang, K.; de la Torre, D.; Funke, L.F.H.; Robertson, W.E.; Christova, Y.; Chia, T.; Schmied, W.H.; Dunkelmann, D.L.; Beránek, V.; et al. Total synthesis of Escherichia coli with a recoded genome. Nature 2019, 569, 514–518. [Google Scholar] [CrossRef]
- Suzuki, T.; Miller, C.; Guo, L.T.; Ho, J.M.L.; Bryson, D.I.; Wang, Y.S.; Liu, D.R.; Söll, D. Crystal structures reveal an elusive functional domain of pyrrolysyl-tRNA synthetase. Nat. Chem. Biol. 2017, 13, 1261–1266. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, T.; Ishii, R.; Fukunaga, R.; Nureki, O.; Yokoyama, S. Crystallization and preliminary X-ray crystallographic analysis of the catalytic domain of pyrrolysyl-tRNA synthetase from the methanogenic archaeon Methanosarcina mazei. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2006, 62, 1031–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herring, S.; Ambrogelly, A.; Gundllapalli, S.; O’Donoghue, P.; Polycarpo, C.R.; Söll, D. The amino-terminal domain of pyrrolysyl-tRNA synthetase is dispensable in vitro but required for in vivo activity. FEBS Lett. 2007, 581, 3197–3203. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, T.; Ishii, R.; Fukunaga, R.; Kobayashi, T.; Sakamoto, K.; Yokoyama, S. Crystallographic Studies on Multiple Conformational States of Active-site Loops in Pyrrolysyl-tRNA Synthetase. J. Mol. Biol. 2008, 378, 634–652. [Google Scholar] [CrossRef]
- Borrel, G.; Parisot, N.; Harris, H.M.B.; Peyretaillade, E.; Gaci, N.; Tottey, W.; Bardot, O.; Raymann, K.; Gribaldo, S.; Peyret, P.; et al. Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genom. 2014, 15, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Kavran, J.M.; Gundllapalli, S.; O′Donoghue, P.; Englert, M.; Söll, D.; Steitz, T.A. Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation. Proc. Natl. Acad. Sci. USA 2007, 104, 11268–11273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takimoto, J.K.; Dellas, N.; Noel, J.P.; Wang, L. Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem. Biol. 2011, 6, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Schneider, S.; Gattner, M.J.; Vrabel, M.; Flügel, V.; López-Carrillo, V.; Prill, S.; Carell, T. Structural insights into incorporation of norbornene amino acids for click modification of proteins. ChemBioChem 2013, 14, 2114–2118. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, T.; Sumida, T.; Ishii, R.; Yokoyama, S. A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 5–15. [Google Scholar] [CrossRef]
- Flügel, V.; Vrabel, M.; Schneider, S. Structural basis for the site-specific incorporation of lysine derivatives into proteins. PLoS ONE 2014, 9, e96198. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.J.; Weber, A.; Pott, M.; Welte, W.; Summerer, D. Structural basis of furan-amino acid recognition by a polyspecific aminoacyl-tRNA-synthetase and its genetic encoding in human cells. ChemBioChem 2014, 15, 1755–1760. [Google Scholar] [CrossRef]
- Guo, L.T.; Wang, Y.S.; Nakamura, A.; Eiler, D.; Kavran, J.M.; Wong, M.; Kiessling, L.L.; Steitz, T.A.; O’Donoghue, P.; Söll, D. Polyspecific pyrrolysyl-tRNA synthetases from directed evolution. Proc. Natl. Acad. Sci. USA 2014, 111, 16724–16729. [Google Scholar] [CrossRef] [Green Version]
- Englert, M.; Nakamura, A.; Wang, Y.S.; Eiler, D.; Söll, D.; Guo, L.T. Probing the active site tryptophan of Staphylococcus aureus thioredoxin with an analog. Nucleic Acids Res. 2015, 43, 11061–11067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.J.; Schmidt, M.J.; Tharp, J.M.; Weber, A.; Koenig, A.L.; Zheng, H.; Gao, J.; Waters, M.L.; Summerer, D.; Liu, W.R. Genetically encoded fluorophenylalanines enable insights into the recognition of lysine trimethylation by an epigenetic reader. Chem. Commun. 2016, 52, 12606–12609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yanagisawa, T.; Kuratani, M.; Seki, E.; Hino, N.; Sakamoto, K.; Yokoyama, S. Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase. Cell Chem. Biol. 2019, 26, 936–949.e13. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.K.; Wang, Y.H.; Weng, J.H.; Kurkute, P.; Li, C.L.; Lee, M.N.; Chen, P.J.; Tseng, H.W.; Tsai, M.D.; Wang, Y.S. Probing the Active Site of Deubiquitinase USP30 with Noncanonical Tryptophan Analogues. Biochemistry 2020, 59, 2205–2209. [Google Scholar] [CrossRef]
- Vatansever, E.C.; Yang, K.S.; Geng, Z.Z.; Qiao, Y.; Li, P.; Xu, S.; Liu, W.R. A Designed, Highly Efficient Pyrrolysyl-tRNA Synthetase Mutant Binds o-Chlorophenylalanine Using Two Halogen Bonds. J. Mol. Biol. 2022, 434, 167534. [Google Scholar] [CrossRef]
- Plass, T.; Milles, S.; Koehler, C.; Schultz, C.; Lemke, E.A. Genetically encoded copper-free click chemistry. Angew. Chem. Int. Ed. Engl. 2011, 50, 3878–3881. [Google Scholar] [CrossRef] [Green Version]
- Plass, T.; Milles, S.; Koehler, C.; Szymański, J.; Mueller, R.; Wiessler, M.; Schultz, C.; Lemke, E.A. Amino acids for Diels-Alder reactions in living cells. Angew. Chem. Int. Ed. Engl. 2012, 51, 4166–4170. [Google Scholar] [CrossRef] [PubMed]
- Borrmann, A.; Milles, S.; Plass, T.; Dommerholt, J.; Verkade, J.M.; Wiessler, M.; Schultz, C.; van Hest, J.C.; van Delft, F.L.; Lemke, E.A. Genetic encoding of a bicyclo[6.1.0]nonyne-charged amino acid enables fast cellular protein imaging by metal-free ligation. ChemBioChem. 2012, 13, 2094–2099. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, T.; Hino, N.; Iraha, F.; Mukai, T.; Sakamoto, K.; Yokoyama, S. Wide-range protein photo-crosslinking achieved by a genetically encoded N(ε)-(benzyloxycarbonyl)lysine derivative with a diazirinyl moiety. Mol. Biosyst. 2012, 8, 1131–1135. [Google Scholar] [CrossRef]
- Nikić, I.; Plass, T.; Schraidt, O.; Szymański, J.; Briggs, J.A.; Schultz, C.; Lemke, E.A. Minimal tags for rapid dual-color live-cell labeling and super-resolution microscopy. Angew. Chem. Int. Ed. Engl. 2014, 53, 2245–2249. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Matsuda, T.; Ohtake, K.; Yanagisawa, T.; Yokoyama, S.; Fujiwara, Y.; Watanabe, T.; Hohsaka, T.; Sakamoto, K. Incorporation of a Doubly Functionalized Synthetic Amino Acid into Proteins for Creating Chemical and Light-Induced Conjugates. Bioconjug. Chem. 2016, 27, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Kita, A.; Hino, N.; Higashi, S.; Hirota, K.; Narumi, R.; Adachi, J.; Takafuji, K.; Ishimoto, K.; Okada, Y.; Sakamoto, K.; et al. Adenovirus vector-based incorporation of a photo-cross-linkable amino acid into proteins in human primary cells and cancerous cell lines. Sci. Rep. 2016, 6, 36946. [Google Scholar] [CrossRef] [Green Version]
- Kato, A.; Kuratani, M.; Yanagisawa, T.; Ohtake, K.; Hayashi, A.; Amano, Y.; Kimura, K.; Yokoyama, S.; Sakamoto, K.; Shiraishi, Y. Extensive Survey of Antibody Invariant Positions for Efficient Chemical Conjugation Using Expanded Genetic Codes. Bioconjug. Chem. 2017, 28, 2099–2108. [Google Scholar] [CrossRef]
- Hoffmann, J.E.; Dziuba, D.; Stein, F.; Schultz, C. A Bifunctional Noncanonical Amino Acid: Synthesis, Expression, and Residue-Specific Proteome-wide Incorporation. Biochemistry 2018, 57, 4747–4752. [Google Scholar] [CrossRef] [Green Version]
- Baumann, T.; Hauf, M.; Richter, F.; Albers, S.; Möglich, A.; Ignatova, Z.; Budisa, N. Computational aminoacyl-tRNA synthetase library design for photocaged tyrosine. Int. J. Mol. Sci. 2019, 20, 2343. [Google Scholar] [CrossRef] [Green Version]
- Gottfried-Lee, I.; Perona, J.J.; Karplus, P.A.; Mehl, R.A.; Cooley, R.B. Structures of Methanomethylophilus alvus Pyrrolysine tRNA-Synthetases Support the Need for De Novo Selections When Altering the Substrate Specificity. ACS Chem. Biol. 2022, 17, 3470–3477. [Google Scholar] [CrossRef]
- Collaborative Computational Project, No. 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994, 50, 760–763. [CrossRef] [PubMed]
- Davis, I.W.; Leaver-Fay, A.; Chen, V.B.; Block, J.N.; Kapral, G.J.; Wang, X.; Murray, L.W.; Arendall, W.B.; Snoeyink, J.; Richardson, J.S.; et al. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007, 35, 375–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eriani, G.; Delarue, M.; Poch, O.; Gangloff, J.; Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 1990, 347, 203–206. [Google Scholar] [CrossRef]
- Cusack, S.; Berthet-Colominas, C.; Härtlein, M.; Nassar, N.; Leberman, R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 Å. Nature 1990, 347, 249–255. [Google Scholar] [CrossRef]
- Ruff, M.; Krishnaswamy, S.; Boeglin, M.; Poterszman, A.; Mitschler, A.; Podjarny, A.; Rees, B.; Thierry, J.C.; Morast, D. Class II aminoacyl transfer RNA synthetases: Crystal structure of yeast apartyl-tRNA synthetase complexed with tRNA(Asp). Science 1991, 252, 1682–1689. [Google Scholar] [CrossRef] [PubMed]
- Kolb, H.C.; Finn, M.G.; Sharpless, K.B. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angew. Chemie-Int. Ed. 2001, 40, 2004–2021. [Google Scholar] [CrossRef]
- Kabsch, W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Kigawa, T.; Yabuki, T.; Matsuda, N.; Matsuda, T.; Tanaka, A.; Yokoyama, S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genom. 2004, 5, 63–68. [Google Scholar] [CrossRef]
- Seki, E.; Matsuda, N.; Yokoyama, S.; Kigawa, T. Cell-free protein synthesis system from Escherichia coli cells cultured at decreased temperatures improves productivity by decreasing DNA template degradation. Anal. Biochem. 2008, 377, 156–161. [Google Scholar] [CrossRef] [PubMed]
- Chumpolkulwong, N.; Hori-Takemoto, C.; Hosaka, T.; Inaoka, T.; Kigawa, T.; Shirouzu, M.; Ochi, K.; Yokoyama, S. Effects of Escherichia coli ribosomal protein S12 mutations on cell-free protein synthesis. Eur. J. Biochem. 2004, 271, 1127–1134. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yanagisawa, T.; Seki, E.; Tanabe, H.; Fujii, Y.; Sakamoto, K.; Yokoyama, S. Crystal Structure of Pyrrolysyl-tRNA Synthetase from a Methanogenic Archaeon ISO4-G1 and Its Structure-Based Engineering for Highly-Productive Cell-Free Genetic Code Expansion with Non-Canonical Amino Acids. Int. J. Mol. Sci. 2023, 24, 6256. https://doi.org/10.3390/ijms24076256
Yanagisawa T, Seki E, Tanabe H, Fujii Y, Sakamoto K, Yokoyama S. Crystal Structure of Pyrrolysyl-tRNA Synthetase from a Methanogenic Archaeon ISO4-G1 and Its Structure-Based Engineering for Highly-Productive Cell-Free Genetic Code Expansion with Non-Canonical Amino Acids. International Journal of Molecular Sciences. 2023; 24(7):6256. https://doi.org/10.3390/ijms24076256
Chicago/Turabian StyleYanagisawa, Tatsuo, Eiko Seki, Hiroaki Tanabe, Yoshifumi Fujii, Kensaku Sakamoto, and Shigeyuki Yokoyama. 2023. "Crystal Structure of Pyrrolysyl-tRNA Synthetase from a Methanogenic Archaeon ISO4-G1 and Its Structure-Based Engineering for Highly-Productive Cell-Free Genetic Code Expansion with Non-Canonical Amino Acids" International Journal of Molecular Sciences 24, no. 7: 6256. https://doi.org/10.3390/ijms24076256