Two Subgroups within the GH43_36 α-l-Arabinofuranosidase Subfamily Hydrolyze Arabinosyl from Either Mono-or Disubstituted Xylosyl Units in Wheat Arabinoxylan
Abstract
:1. Introduction
2. Results
2.1. Structure-Based Phylogenetic Analysis of GH43_36 Reveals Further Subdivision with Distinct GH43_36a and GH43_36b Subgroups
2.2. Production, Purification, and General Characteristics of TpABF43_36b
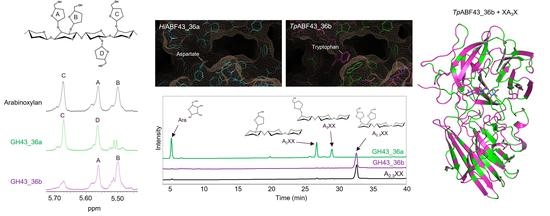
2.3. GH43_36a Cleaves α-(1→3)-Ara from Disubstituted Xyls (ABF-d3) and GH43_36b Cleaves Ara from Monosubstituted Xyls
2.4. Main and Side Activities of TpABF43_36b and HiABF43_36a on AX and AXOS
2.5. HiABF43_36a Cleaves α-(1→2 or 3)-Ara from Xyls Disubstituted on the Non-Reducing End and α-(1→2 and 3)-Ara Monosubstituted on the Second Xyl from the Non-Reducing End
2.6. Rate and Synergy of the GH43_36, GH51, and GH62 Enzymes used in this Study
2.6.1. Rate of TpABF43_36b, HiABF43_36, MgABF51, and PoABF62 Enzymes on AX
2.6.2. Rate of TpABF43_36b, HiABF43_36, MgABF51, and PoABF62 on Specific AXOS
2.6.3. Synergy of TpABF43_36b, HiABF43_36, MgABF51, and PoABF62 on AX
3. Discussion
4. Materials and Methods
4.1. Materials
4.1.1. Chemicals and Substrate
4.1.2. Enzymes
4.2. Enzyme Characterization
4.2.1. Phylogenetic Analysis of GH43_36 from Ascomycetes
4.2.2. Enzyme Incubations
4.2.3. Analysis of ABF Activity with 1H-NMR
4.2.4. Analysis of Reaction Products with HPAEC-PAD
4.2.5. Analysis of Reaction Products with MALDI-TOF MS
4.2.6. Analysis of Reducing End Groups with PAHBAH Assay
4.2.7. AZCL-Xylanase Activity
4.2.8. Production of AXOS Monosubstituted on the Second Xylosyl from the Non-Reducing End
4.2.9. Proteomics: Protein ID
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lynd, L. Overview and evaluation of fuel ethanol from cellulosic biomass: Technology, economics, the environment, and policy. Annu. Rev. Energy Environ. 1996, 21, 403–465. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Chatzifragkou, A.; Kosik, O.; Gibson, G.; Lovegrove, A.; Shewry, P.; Charalampopoulos, D. Evaluation of the prebiotic potential of arabinoxylans extracted from wheat distillers’ dried grains with solubles (DDGS) and in-process samples. Appl. Microbiol. Biotechnol. 2018, 102, 7577–7587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramseyer, D.; Bettge, A.; Morris, C. Distribution of total, water-unextractable, and water-extractable arabinoxylans in wheat flour mill streams. Cereal Chem. 2011, 88, 209–216. [Google Scholar] [CrossRef]
- Saulnier, L.; Thibault, J. Ferulic acid and diferulic acids as components of sugar-beet pectins and maize bran heteroxylans. J. Sci. Food Agric. 1999, 79, 396–402. [Google Scholar] [CrossRef]
- Saulnier, L.; Guillon, F.; Chateigner-Boutin, A. Cell wall deposition and metabolism in wheat grain. J. Cereal Sci. 2012, 56, 91–108. [Google Scholar] [CrossRef]
- Ebringerova, A.; Hromadkova, Z.; Heinze, T. Hemicellulose. In Polysaccharides I: Structure, Characterization and Use; Springer: Berlin/Heidelberg, Germany, 2005; Volume 186, pp. 1–67. [Google Scholar]
- Barron, C.; Robert, P.; Guillon, F.; Saulnier, L.; Rouau, X. Structural heterogeneity of wheat arabinoxylans revealed by Raman spectroscopy. Carbohydr. Res. 2006, 341, 1186–1191. [Google Scholar] [CrossRef]
- Sørensen, H.; Jørgensen, C.; Hansen, C.; Jørgensen, C.; Pedersen, S.; Meyer, A. A novel GH43 α-l-arabinofuranosidase from Humicola insolens: Mode of action and synergy with GH51 α-l-arabinofuranosidases on wheat arabinoxylan. Appl. Microbiol. Biotechnol. 2006, 73, 850–861. [Google Scholar] [CrossRef]
- Drula, E.; Garron, M.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The carbohydrate-active enzyme database: Functions and literature. Nucleic Acids Res. 2022, 50, 571–577. [Google Scholar] [CrossRef]
- Lagaert, S.; Pollet, A.; Courtin, C.; Volckaert, G. β-xylosidases and α-l-arabinofuranosidases: Accessory enzymes for arabinoxylan degradation. Biotechnol. Adv. 2014, 32, 316–332. [Google Scholar] [CrossRef]
- Koutaniemi, S.; Tenkanen, M. Action of three GH51 and one GH54 α-arabinofuranosidases on internally and terminally located arabinofuranosyl branches. J. Biotechnol. 2016, 229, 22–30. [Google Scholar] [CrossRef]
- McKee, L.; Peña, M.; Rogowski, A.; Jackson, A.; Lewis, R.; York, W.; Krogh, K.; Viksø-Nielsen, A.; Skjøt, M.; Gilbert, H.; et al. Introducing endo-xylanase activity into an exo-acting arabinofuranosidase that targets side chains. Proc. Natl. Acad. Sci. USA 2012, 109, 6537–6542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Laere, K.M.J.; Beldman, G.; Voragen, A. A new arabinofuranohydrolase from Bifidobacterium adolescentis able to remove arabinosyl residues from double-substituted xylose units in arabinoxylan. Appl. Microbiol. Biotechnol. 1997, 47, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Orita, T.; Sakka, M.; Kimura, T.; Sakka, K. Characterization of Ruminiclostridium josui arabinoxylan arabinofuranohydrolase, RjAxh43B, and RjAxh43B-containing xylanolytic complex. Enzyme Microb. Technol. 2017, 104, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Pouvreau, L.; Joosten, R.; Hinz, S.; Gruppen, H.; Schols, H. Chrysosporium lucknowense C1 arabinofuranosidases are selective in releasing arabinose from either single or double substituted xylose residues in arabinoxylans. Enzyme Microb. Technol. 2011, 48, 397–403. [Google Scholar] [CrossRef]
- Komeno, M.; Yoshihara, Y.; Kawasaki, J.; Nabeshima, W.; Maeda, K.; Sasaki, Y.; Fujita, K.; Ashida, H. Two α-l-arabinofuranosidases from Bifidobacterium longum subsp. longum are involved in arabinoxylan utilization. Appl. Microbiol. Biotechnol. 2022, 106, 1957–1965. [Google Scholar] [CrossRef]
- Shinozaki, A.; Hosokawa, S.; Nakazawa, M.; Ueda, M.; Sakamoto, T. Identification and characterization of three Penicillium chrysogenum α-l-arabinofuranosidases (PcABF43B, PcABF51C, and AFQ1) with different specificities toward arabino-oligosaccharides. Enzyme Microb. Technol. 2015, 73–74, 65–71. [Google Scholar] [CrossRef]
- Yan, Q.; Tang, L.; Yang, S.; Zhou, P.; Zhang, S.; Jiang, Z. Purification and characterization of a novel thermostable α-l-arabinofuranosidase (α-l-AFase) from Chaetomium sp. Process Biochem. 2012, 47, 472–478. [Google Scholar] [CrossRef]
- Kühnel, S.; Westphal, Y.; Hinz, S.; Schols, H.; Gruppen, H. Mode of action of Chrysosporium lucknowense C1 α-l-arabinohydrolases. Bioresour. Technol. 2011, 102, 1636–1643. [Google Scholar] [CrossRef]
- Yang, X.; Shi, P.; Ma, R.; Luo, H.; Huang, H.; Yang, P.; Yao, B. A new GH43 α-arabinofuranosidase from Humicola insolens Y1: Biochemical characterization and synergistic action with a xylanase on xylan degradation. Appl. Biochem. Biotechnol. 2014, 175, 1960–1970. [Google Scholar] [CrossRef]
- Shinozaki, A.; Kawakami, T.; Hosokawa, S.; Sakamoto, T. A novel GH43 α-l-arabinofuranosidase of Penicillium chrysogenum that preferentially degrades single-substituted arabinosyl side chains in arabinan. Enzyme Microb. Technol. 2014, 58–59, 80–86. [Google Scholar] [CrossRef]
- Ravanal, M.; Alegría-Arcos, M.; Gonzalez-Nilo, F.; Eyzaguirre, J. Penicillium purpurogenum produces two GH family 43 enzymes with β-xylosidase activity, one monofunctional and the other bifunctional: Biochemical and structural analyses explain the difference. Arch. Biochem. Biophys. 2013, 540, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, S.; Zhang, T.; Xian, L.; Liao, L.; Liu, J.; Feng, J. Genome sequencing and analysis of Talaromyces pinophilus provide insights into biotechnological applications. Sci. Rep. 2017, 7, 490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalboege, H.; Heldt-Hansen, H.P. A novel method for efficient expression cloning of fungal enzyme genes. Mol. Gen. Genet. 1994, 243, 253–260. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Kouzounis, D.; Kabel, M.; De Vries, R. GH10 and GH11 endoxylanases in Penicillium subrubescens: Comparative characterization and synergy with GH51, GH54, GH62 α-l-arabinofuranosidases from the same fungus. New Biotechnol. 2022, 70, 84–92. [Google Scholar] [CrossRef]
- Mewis, K.; Lenfant, N.; Lombard, V.; Henrissat, B. Dividing the large glycoside hydrolase family 43 into subfamilies: A motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 2016, 82, 1686–1692. [Google Scholar] [CrossRef] [Green Version]
- Huy, N.; Thayumanavan, P.; Kwon, T.; Park, S. Characterization of a recombinant bifunctional xylosidase/arabinofuranosidase from Phanerochaete chrysosporium. J. Biosci. Bioeng. 2013, 116, 152–159. [Google Scholar] [CrossRef]
- Carvalho, D.; Carli, S.; Meleiro, L.; Rosa, J.; Oliveira, A.; Jorge, J.; Furriel, R. A halotolerant bifunctional β-xylosidase/α-l-arabinofuranosidase from Colletotrichum graminicola: Purification and biochemical characterization. Int. J. Biol. Macromol. 2018, 114, 741–750. [Google Scholar] [CrossRef]
- Pollet, A.; Delcour, J.; Courtin, C. Structural determinants of the substrate specificities of xylanases from different glycoside hydrolase families. Crit. Rev. Biotechnol. 2010, 30, 176–191. [Google Scholar] [CrossRef]
- Coconi Linares, N.; Li, X.; Dilokpimol, A.; Vries, R. Comparative characterization of nine novel GH51, GH54 and GH62 α-l-arabinofuranosidases from Penicillium subrubescens. FEBS Lett. 2022, 596, 360–368. [Google Scholar] [CrossRef]
- Wang, W.; Mai-Gisondi, G.; Stogios, P.; Kaur, A.; Xu, X.; Cui, H.; Turunen, O.; Savchenko, A.; Master, E. Elucidation of the molecular basis for arabinoxylan-debranching activity of a thermostable family GH62 α-l-arabinofuranosidase from Streptomyces thermoviolaceus. Appl. Environ. Microbiol. 2014, 80, 5317–5329. [Google Scholar] [CrossRef]
- Souza, T.; Santos, C.; Souza, A.; Oldiges, D.; Ruller, R.; Prade, R.; Squina, F.; Murakami, M. Structure of a novel thermostable GH51 α-l-arabinofuranosidase from Thermotoga petrophila RKU-1. Protein Sci. 2011, 20, 1632–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkens, C.; Andersen, S.; Dumon, C.; Berrin, J.; Svensson, B. GH62 arabinofuranosidases: Structure, function and applications. Biotechnol. Adv. 2017, 35, 792–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szabo, L.; Jamal, S.; Xie, H.; Charnock, S.; Bolam, D.; Gilbert, H.; Davies, G. Structure of a family 15 carbohydrate-binding module in complex with xylopentaose. Evidence that xylan binds in an approximate 3-fold helical conformation. J. Biol. Chem. 2001, 276, 49061–49065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.; Dvorkin, M.; Kulikov, A.; Lesin, V.; Nikolenko, S.; Pham, S.; Prjibelski, A.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ter-Hovhannisyan, V.; Lomsadze, A.; Chernoff, Y.; Borodovsky, M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008, 18, 1979–1990. [Google Scholar] [CrossRef] [Green Version]
- Stanke, M.; Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 2003, 19, 215–225. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Higgins, D. The clustal omega multiple alignment package. Methods Mol. Biol. 2020, 2231, 3–16. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, 293–296. [Google Scholar] [CrossRef]












| Class | Enzyme | Organism | Selectivity | Source |
|---|---|---|---|---|
| GH43_36 | HiABF43_36a 1 | Humicola insolens | ABF-d3 | [8] |
| Abn7 | Chrysosporium lucknowense | [15] | ||
| GH43_21 | Abf43B | Penicillium chrysogenum | ABF-m2,3 | [17] |
| GH43_26 | CsAra | Chaetomium sp. | ABF-m2,3 | [18] |
| Abn4 | Chrysosporium lucknowense | [19] | ||
| HiAbf43 | Humicola insolens | [20] | ||
| PcABF43A | Penicillium chrysogenum | [21] | ||
| GH43_29 | TpAra43_29 | Talaromyces purpureogenus | ABF-m2,3 | [22] |
| GH51 | MgABF51 1 | Meripilus giganteus | ABF-m2,3 ABF-d3 2 | [8,11] |
| GH62 | PoABF62 1 | Penicillium oxalicum | ABF-m2,3 | - |
| Enzyme | ABF | Xylanase | AXOS | |||||
|---|---|---|---|---|---|---|---|---|
| Selectivity | Rate (AX) | Selectivity | Rate (AX) | A2,3XX | A2XX | XA2XX | XA3XX | |
| HiABF43_36a | ABF-d3 | ++ | -- | -- | +++ | -- | +/− | +/− |
| TpABF43_36b | ABF-m2,3 | + | Endo | +/− | -- | +/− | + | +/− |
| MgABF51 | ABF-2,3 ABF-d3 * | +++ | -- | -- | + | +++ | +++ | +++ |
| PoABF62 | ABF-m2,3 | +++ | Exo | - | - | ++ | +++ | +++ |
| No. | Enzyme Name | Time (h) | Enzyme (μg mL−1) | Substrate Type | Substrate (μg mL−1) | Analysis Method |
|---|---|---|---|---|---|---|
| 1 | MgABF51 | 1, 24 | 1, 50 | AX | 1000 | 1H-NMR, HPLC, MS |
| PoABF62 | ||||||
| HiABF43_36a | ||||||
| TpABF43_36b | ||||||
| 2 | MgABF51 | 24 | 0 to 25 | A2XX, XA3XX, A2,3XX, XA2XX/XA3XX | 100 | HPLC |
| PoABF62 | ||||||
| HiABF43_36a | ||||||
| TpABF43_36b | ||||||
| 3 | HiABF43_36a | 1 | 0 to 50 | AX | 1000 | PAHBAH, HPLC |
| MgABF51 | ||||||
| PoABF62 | ||||||
| HiABF43_36a + MgABF51 | ||||||
| HiABF43_36a + PoABF62 | ||||||
| MgABF51, PoABF62 | ||||||
| 4 | TpABF43_36b | 1, 23 | 0 to 50 | AX | 1000 | PAHBAH, HPLC |
| TpABF43_36b + HiABF43_36a | ||||||
| TpABF43_36b + MgABF51 | ||||||
| 5 | TpABF43_36b | 1 | 0 to 25 | AB, RG | 1000 | PAHBAH |
| 6 | TpABF43_36b | 1 | 0 to 25 | Corn fiber | 30.000 | PAHBAH, HPLC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leschonski, K.P.; Kaasgaard, S.G.; Spodsberg, N.; Krogh, K.B.R.M.; Kabel, M.A. Two Subgroups within the GH43_36 α-l-Arabinofuranosidase Subfamily Hydrolyze Arabinosyl from Either Mono-or Disubstituted Xylosyl Units in Wheat Arabinoxylan. Int. J. Mol. Sci. 2022, 23, 13790. https://doi.org/10.3390/ijms232213790
Leschonski KP, Kaasgaard SG, Spodsberg N, Krogh KBRM, Kabel MA. Two Subgroups within the GH43_36 α-l-Arabinofuranosidase Subfamily Hydrolyze Arabinosyl from Either Mono-or Disubstituted Xylosyl Units in Wheat Arabinoxylan. International Journal of Molecular Sciences. 2022; 23(22):13790. https://doi.org/10.3390/ijms232213790
Chicago/Turabian StyleLeschonski, Kai P., Svend G. Kaasgaard, Nikolaj Spodsberg, Kristian B. R. M. Krogh, and Mirjam A. Kabel. 2022. "Two Subgroups within the GH43_36 α-l-Arabinofuranosidase Subfamily Hydrolyze Arabinosyl from Either Mono-or Disubstituted Xylosyl Units in Wheat Arabinoxylan" International Journal of Molecular Sciences 23, no. 22: 13790. https://doi.org/10.3390/ijms232213790